浙江农业学报 ›› 2026, Vol. 38 ›› Issue (1): 35-53.DOI: 10.3969/j.issn.1004-1524.20250081
藜麦GRAS基因家族的鉴定及其在生殖发育中的调控功能
杨炀a( ), 张帅a, 董陈文华a,b, 曾孟琼a, 林春a,b, 毛自朝a,b,*(
), 张帅a, 董陈文华a,b, 曾孟琼a, 林春a,b, 毛自朝a,b,*( )
)
- 云南农业大学 a. 农业与生物技术学院;b. 特色小宗作物研究中心,云南 昆明 650201
-
收稿日期:2025-02-07出版日期:2026-01-25发布日期:2026-02-11 -
作者简介:毛自朝,E-mail:zmao@ynau.edu.cn
杨炀,研究方向为生物化学与分子生物学。E-mail:Yyang_ynau@163.com -
通讯作者:毛自朝 -
基金资助:国家自然科学基金(32260482)
Identification of GRAS gene family members and their regulatory roles in reproductive development in Chenopodium quinoa
YANG Yanga( ), ZHANG Shuaia, DONG Chenwenhuaa,b, ZENG Mengqionga, LIN Chuna,b, MAO Zichaoa,b,*(
), ZHANG Shuaia, DONG Chenwenhuaa,b, ZENG Mengqionga, LIN Chuna,b, MAO Zichaoa,b,*( )
)
- College of Agriculture and Biotechnology; b. Research Center of Featured Minor Crops, Yunnan Agricultural University, Kunming 650201, China
-
Received:2025-02-07Online:2026-01-25Published:2026-02-11 -
Contact:MAO Zichao
摘要:
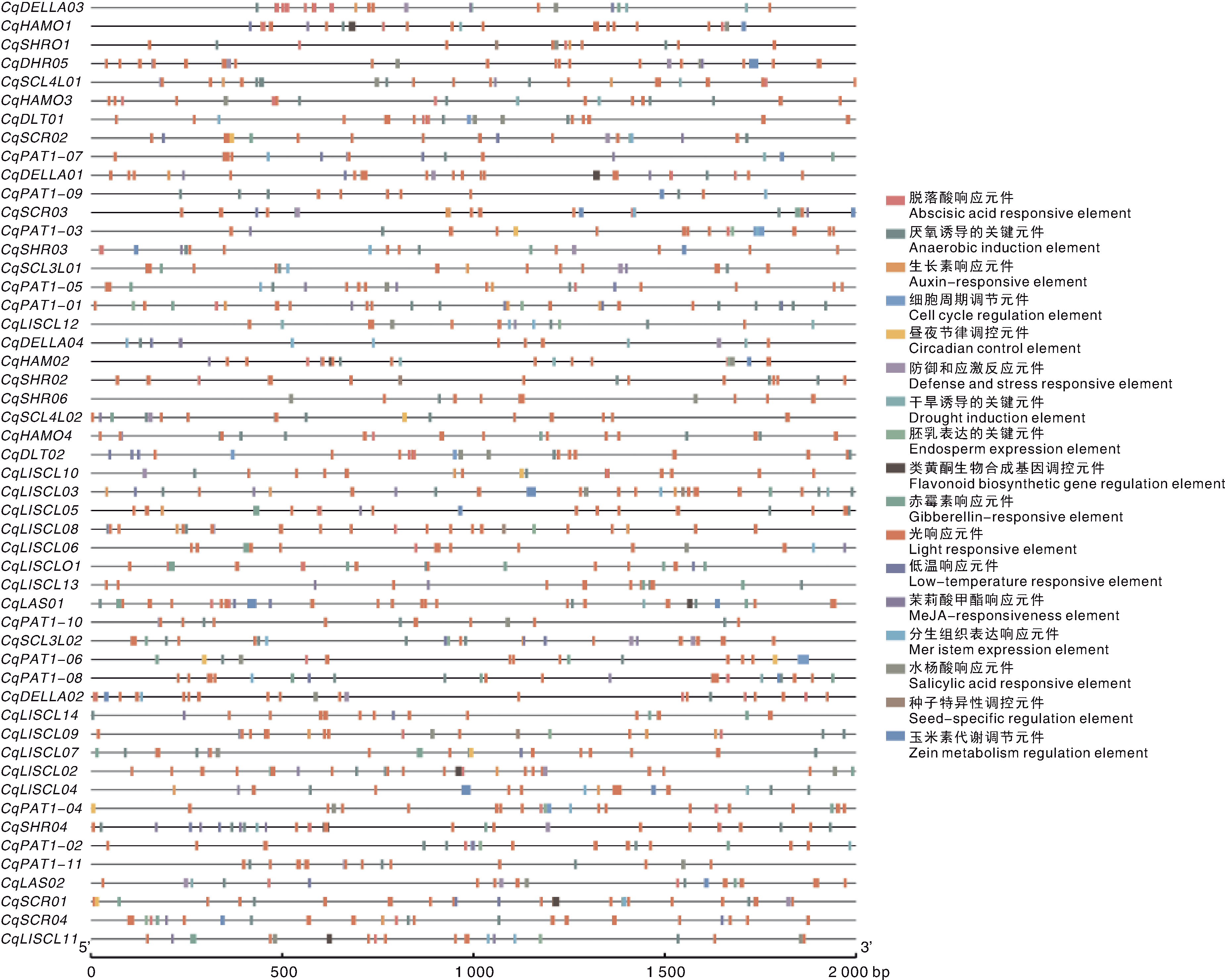
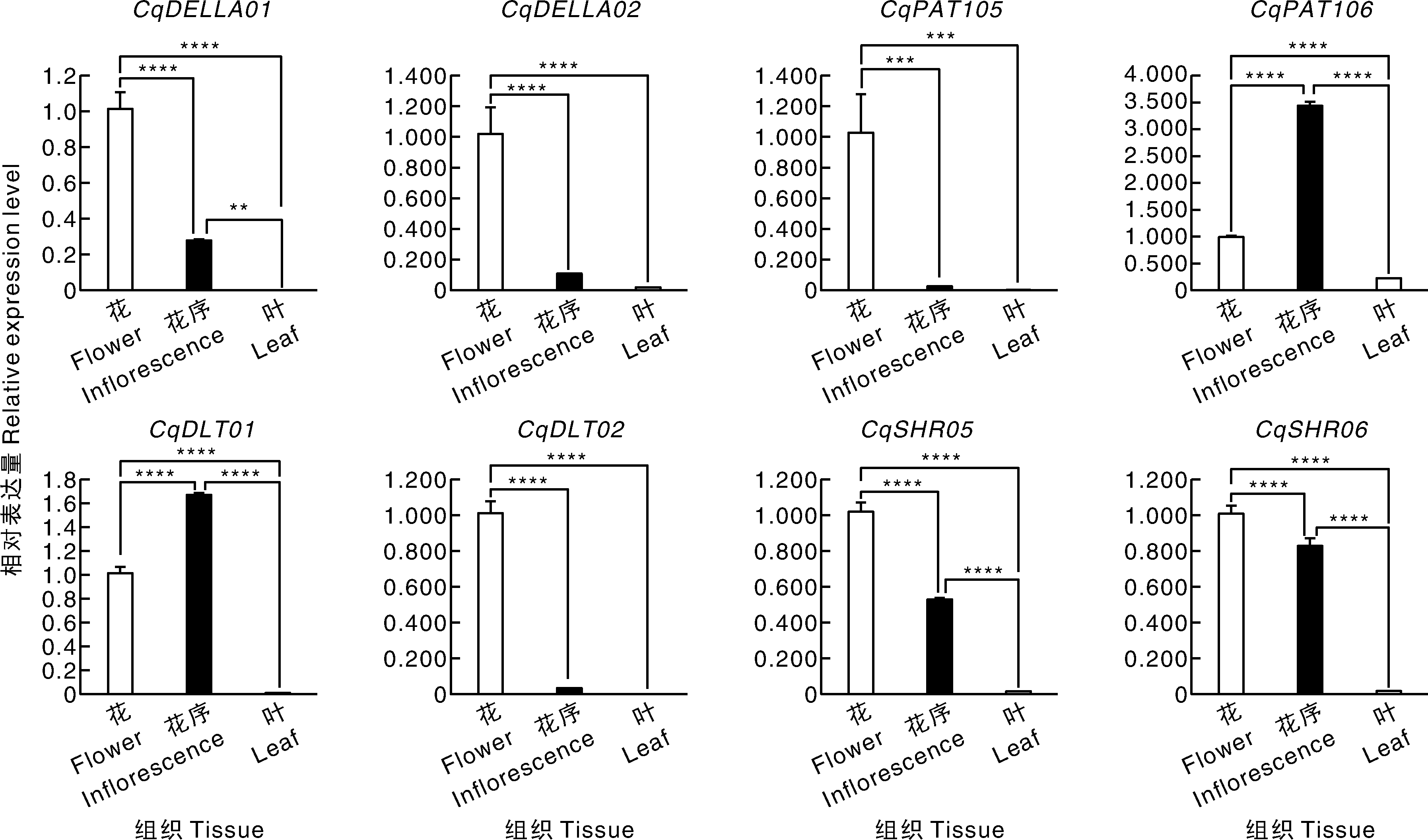
GRAS基因家族在植物生长发育与逆境响应中具有重要调控功能,但目前尚未见其在藜麦(Chenopodium quinoa)生殖发育中的相关报道。本研究基于最新藜麦基因组数据,系统鉴定并分析藜麦GRAS家族(CqGRAS)成员,重点解析其基因结构、启动子顺式作用元件,以及在营养与生殖生长阶段的表达模式与调控机制。同时,将CqGRAS成员与拟南芥、藜麦二倍体祖先Chenopodium watsonii(A基因组)和Chenopodium suecicum(B基因组)的GRAS基因进行对比分析。结果共鉴定到51个CqGRAS基因,这些基因普遍内含子数量较少,且与拟南芥及二倍体藜属物种GRAS基因具有较高同源性。启动子分析表明,该家族基因富含响应植物激素(如赤霉素、脱落酸、乙烯和茉莉酸)、生长发育与逆境胁迫的顺式作用元件。系统进化分析将CqGRAS家族划分为10个亚家族,其中HAM(CqHAM01)、PAT1(CqPAT1-06/07/08)、DELLA(CqDELLA01/02)、DLT(CqDLT01/02)和SHR(CqSHR05/06)在花序和发育种子中表达水平较高。加权基因共表达网络分析提示,这些与生殖发育相关的CqGRAS基因可能通过整合光信号与激素信号通路,调控藜麦花和种子的生长发育。本研究为阐明GRAS家族在藜麦生殖发育中的功能提供了新见解,并为深入解析其分子机制与育种应用奠定了基础。
中图分类号:
引用本文
杨炀, 张帅, 董陈文华, 曾孟琼, 林春, 毛自朝. 藜麦GRAS基因家族的鉴定及其在生殖发育中的调控功能[J]. 浙江农业学报, 2026, 38(1): 35-53.
YANG Yang, ZHANG Shuai, DONG Chenwenhua, ZENG Mengqiong, LIN Chun, MAO Zichao. Identification of GRAS gene family members and their regulatory roles in reproductive development in Chenopodium quinoa[J]. Acta Agriculturae Zhejiangensis, 2026, 38(1): 35-53.
| 引物名称primer name | 引物序列(5'→3') Primer sequence(5'→3') | 产物大小/bp Product length/bp |
|---|---|---|
| 1-CqDELLA01-F2 | CGAAAGACACGAGCCACTAATAAA | 169 |
| 1-CqDELLA01-R2 | CCACCCTAGAGTCAAGCACCC | |
| 2-CqDELLA02-F2 | ACTTCTAGTTCTCAATCTTTTGTTTTC | 194 |
| 2-CqDELLA02-R2 | AAAGTGTCTAGCACCCCCAG | |
| 3-CqPAT1-05-F2 | CTTTCCCTTAGCAATGACAACA | 105 |
| 3-CqPAT1-05-R2 | AATGTTTAGGAAGGATGGTCGT | |
| 4-CqPAT1-06-F | AGGACTACTTCGGTGTTACTCTG | 103 |
| 4-CqPAT1-06-R | CCAAGCTGAAGCCGAGATCAA | |
| 5-CqDLT01-F2 | GGTATCAGGAGGGACGAAAG | 166 |
| 5-CqDLT01-R2 | CAGCAGCATCAACAGAACCC | |
| 6-CqDLT02-F2 | CATGGATTAGTCTTTGATTTTCGAT | 175 |
| 6-CqDLT02-R2 | CATGCCATAAGTAGATCAAATAACAA | |
| 7-CqSHR05-F | GGCGTATAGTGAGGAGGTTTGTG | 109 |
| 7-CqSHR05-R | TATTCCGGCAGTCGGAGAC | |
| 8-CqSHR06-F | CGGAGAGGAGAGAGAATGCC | 137 |
| 8-CqSHR06-R | TGCCATCGACCACCCTTCT | |
| CqActin-F2 | TTGGAATCTCGCCGCCAAA | 132 |
| CqActin-R2 | CCAGGTCCACCAACAATCCA |
表1 qRT-PCR引物序列
Table 1 Sequences of qRT-PCR primers
| 引物名称primer name | 引物序列(5'→3') Primer sequence(5'→3') | 产物大小/bp Product length/bp |
|---|---|---|
| 1-CqDELLA01-F2 | CGAAAGACACGAGCCACTAATAAA | 169 |
| 1-CqDELLA01-R2 | CCACCCTAGAGTCAAGCACCC | |
| 2-CqDELLA02-F2 | ACTTCTAGTTCTCAATCTTTTGTTTTC | 194 |
| 2-CqDELLA02-R2 | AAAGTGTCTAGCACCCCCAG | |
| 3-CqPAT1-05-F2 | CTTTCCCTTAGCAATGACAACA | 105 |
| 3-CqPAT1-05-R2 | AATGTTTAGGAAGGATGGTCGT | |
| 4-CqPAT1-06-F | AGGACTACTTCGGTGTTACTCTG | 103 |
| 4-CqPAT1-06-R | CCAAGCTGAAGCCGAGATCAA | |
| 5-CqDLT01-F2 | GGTATCAGGAGGGACGAAAG | 166 |
| 5-CqDLT01-R2 | CAGCAGCATCAACAGAACCC | |
| 6-CqDLT02-F2 | CATGGATTAGTCTTTGATTTTCGAT | 175 |
| 6-CqDLT02-R2 | CATGCCATAAGTAGATCAAATAACAA | |
| 7-CqSHR05-F | GGCGTATAGTGAGGAGGTTTGTG | 109 |
| 7-CqSHR05-R | TATTCCGGCAGTCGGAGAC | |
| 8-CqSHR06-F | CGGAGAGGAGAGAGAATGCC | 137 |
| 8-CqSHR06-R | TGCCATCGACCACCCTTCT | |
| CqActin-F2 | TTGGAATCTCGCCGCCAAA | 132 |
| CqActin-R2 | CCAGGTCCACCAACAATCCA |
| 基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 氨基酸长度/aa Amino acid length/aa | 蛋白质分子质量/u Protein molecular weight/u | 等电点 Isoelectric point | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| CqDELLA01 | CQ012679 | Cq8B | 459 | 50 811.86 | 5.51 | 溶酶体Cytolysosome |
| CqDELLA02 | CQ040570 | Cq8A | 474 | 52 509.87 | 5.66 | 溶酶体Cytolysosome |
| CqDELLA03 | CQ001679 | Cq6B | 617 | 67 367.76 | 4.90 | 细胞核Nuclear |
| CqDELLA04 | CQ026984 | Cq6A | 618 | 67 500.87 | 4.90 | 细胞核Nuclear |
| CqDLT01 | CQ004440 | Cq5B | 648 | 73 031.86 | 5.77 | 叶绿体Chloroplast |
| CqDLT02 | CQ029757 | Cq5A | 657 | 73 968.04 | 5.47 | 叶绿体Chloroplast |
| CqHAM01 | CQ002321 | Cq6B | 610 | 67 095.88 | 6.46 | 细胞核Nuclear |
| CqHAM02 | CQ027701 | Cq6A | 602 | 66 232.26 | 6.82 | 细胞核Nuclear |
| CqHAM03 | CQ003474 | Cq5B | 785 | 85 801.61 | 6.15 | 细胞核Nuclear |
| CqHAM04 | CQ028875 | Cq5A | 781 | 85 891.76 | 6.12 | 细胞核Nuclear |
| CqLAS01 | CQ033480 | Cq9B | 490 | 55 803.58 | 7.02 | 细胞核Nuclear |
| CqLAS02 | CQ051297 | Cq9A | 484 | 54 848.37 | 6.76 | 细胞核Nuclear |
| CqLISCL01 | CQ032605 | Cq9B | 350 | 41 049.14 | 8.10 | 溶酶体Cytolysosome |
| CqLISCL02 | CQ041202 | Cq7A | 725 | 82 353.95 | 4.95 | 细胞核Nuclear |
| CqLISCL03 | CQ032598 | Cq9B | 689 | 78 590.85 | 5.18 | 细胞核Nuclear |
| CqLISCL04 | CQ041204 | Cq7A | 619 | 70 272.91 | 6.29 | 细胞核Nuclear |
| CqLISCL05 | CQ032601 | Cq9B | 679 | 76 337.79 | 5.60 | 细胞核Nuclear |
| CqLISCL06 | CQ032604 | Cq9B | 548 | 62 438.96 | 7.94 | 溶酶体Cytolysosome |
| CqLISCL07 | CQ041201 | Cq7A | 662 | 74 879.34 | 6.61 | 细胞核Nuclear |
| CqLISCL08 | CQ032603 | Cq9B | 667 | 75 299.73 | 5.23 | 细胞核Nuclear |
| CqLISCL09 | CQ041200 | Cq7A | 686 | 77 311.71 | 4.87 | 细胞核Nuclear |
| CqLISCL10 | CQ030634 | Cq5A | 760 | 86 162.35 | 5.50 | 细胞核Nuclear |
| CqLISCL11 | CQ055311 | Cq2A | 780 | 87 684.22 | 7.07 | 溶酶体Cytolysosome |
| CqLISCL12 | CQ025002 | Cq1B | 818 | 91 640.62 | 7.49 | 细胞核Nuclear |
| CqLISCL13 | CQ032609 | Cq9B | 756 | 84 422.57 | 5.55 | 细胞核Nuclear |
| CqLISCL14 | CQ041197 | Cq7A | 756 | 84 533.61 | 5.81 | 细胞核Nuclear |
| CqPAT1-01 | CQ024284 | Cq1B | 545 | 61 454.56 | 4.73 | 叶绿体Chloroplast |
| CqPAT1-02 | CQ048458 | Cq1A | 426 | 48 153.43 | 5.27 | 溶酶体Cytolysosome |
| CqPAT1-03 | CQ016853 | Cq3B | 639 | 70 122.51 | 5.46 | 细胞核Nuclear |
| CqPAT1-04 | CQ044170 | Cq3A | 767 | 84 245.91 | 6.39 | 细胞核Nuclear |
| CqPAT1-05 | CQ022180 | Cq4B | 635 | 70 854.40 | 8.16 | 叶绿体Chloroplast |
| CqPAT1-06 | CQ037875 | Cq4A | 634 | 70 854.39 | 7.57 | 叶绿体Chloroplast |
| CqPAT1-07 | CQ010139 | Cq8B | 544 | 61 079.89 | 6.33 | 细胞核Nuclear |
| CqPAT1-08 | CQ038026 | Cq8A | 544 | 61 110.88 | 6.51 | 细胞核Nuclear |
| CqPAT1-09 | CQ013243 | Cq2B | 466 | 52 280.60 | 5.82 | 叶绿体Chloroplast |
| CqPAT1-10 | CQ034654 | Cq9B | 541 | 60 293.76 | 6.56 | 细胞核Nuclear |
| CqPAT1-11 | CQ050106 | Cq9A | 588 | 65 884.16 | 6.69 | 叶绿体Chloroplast |
| CqSCL3L01 | CQ019876 | Cq4B | 454 | 50 710.01 | 6.51 | 细胞核Nuclear |
| CqSCL3L02 | CQ035148 | Cq4A | 454 | 50 837.30 | 6.65 | 细胞核Nuclear |
| CqSCL4L01 | CQ003361 | Cq6B | 557 | 60 789.24 | 4.38 | 细胞核Nuclear |
| CqSCL4L02 | CQ028754 | Cq6A | 610 | 67 157.51 | 4.74 | 细胞核Nuclear |
| CqSCR01 | CQ052618 | Cq9A | 872 | 95 159.47 | 6.39 | 细胞核Nuclear |
| CqSCR02 | CQ007129 | Cq7B | 861 | 93 906.00 | 6.40 | 细胞核Nuclear |
| CqSCR03 | CQ014997 | Cq2B | 396 | 43 813.31 | 6.99 | 叶绿体Chloroplast |
| CqSCR04 | CQ054542 | Cq2A | 396 | 43 859.22 | 7.32 | 细胞核Nuclear |
| CqSHR01 | CQ003291 | Cq6B | 475 | 53 457.35 | 5.88 | 线粒体、细胞骨架 Mitochondrion, cytoskeleton |
| CqSHR02 | CQ028690 | Cq6A | 389 | 43 994.11 | 5.05 | 细胞骨架Cytoskeleton |
| CqSHR03 | CQ018831 | Cq3B | 439 | 50 575.84 | 6.38 | 叶绿体Chloroplast |
| CqSHR04 | CQ044974 | Cq3A | 440 | 50 599.98 | 6.73 | 叶绿体Chloroplast |
| CqSHR05 | CQ003315 | Cq6B | 472 | 53 064.48 | 5.94 | 细胞核Nuclear |
| CqSHR06 | CQ028714 | Cq6A | 467 | 52 415.84 | 5.86 | 细胞核Nuclear |
表2 藜麦GRAS基因家族信息
Table 2 Information of GRAS gene family in quinoa
| 基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 氨基酸长度/aa Amino acid length/aa | 蛋白质分子质量/u Protein molecular weight/u | 等电点 Isoelectric point | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| CqDELLA01 | CQ012679 | Cq8B | 459 | 50 811.86 | 5.51 | 溶酶体Cytolysosome |
| CqDELLA02 | CQ040570 | Cq8A | 474 | 52 509.87 | 5.66 | 溶酶体Cytolysosome |
| CqDELLA03 | CQ001679 | Cq6B | 617 | 67 367.76 | 4.90 | 细胞核Nuclear |
| CqDELLA04 | CQ026984 | Cq6A | 618 | 67 500.87 | 4.90 | 细胞核Nuclear |
| CqDLT01 | CQ004440 | Cq5B | 648 | 73 031.86 | 5.77 | 叶绿体Chloroplast |
| CqDLT02 | CQ029757 | Cq5A | 657 | 73 968.04 | 5.47 | 叶绿体Chloroplast |
| CqHAM01 | CQ002321 | Cq6B | 610 | 67 095.88 | 6.46 | 细胞核Nuclear |
| CqHAM02 | CQ027701 | Cq6A | 602 | 66 232.26 | 6.82 | 细胞核Nuclear |
| CqHAM03 | CQ003474 | Cq5B | 785 | 85 801.61 | 6.15 | 细胞核Nuclear |
| CqHAM04 | CQ028875 | Cq5A | 781 | 85 891.76 | 6.12 | 细胞核Nuclear |
| CqLAS01 | CQ033480 | Cq9B | 490 | 55 803.58 | 7.02 | 细胞核Nuclear |
| CqLAS02 | CQ051297 | Cq9A | 484 | 54 848.37 | 6.76 | 细胞核Nuclear |
| CqLISCL01 | CQ032605 | Cq9B | 350 | 41 049.14 | 8.10 | 溶酶体Cytolysosome |
| CqLISCL02 | CQ041202 | Cq7A | 725 | 82 353.95 | 4.95 | 细胞核Nuclear |
| CqLISCL03 | CQ032598 | Cq9B | 689 | 78 590.85 | 5.18 | 细胞核Nuclear |
| CqLISCL04 | CQ041204 | Cq7A | 619 | 70 272.91 | 6.29 | 细胞核Nuclear |
| CqLISCL05 | CQ032601 | Cq9B | 679 | 76 337.79 | 5.60 | 细胞核Nuclear |
| CqLISCL06 | CQ032604 | Cq9B | 548 | 62 438.96 | 7.94 | 溶酶体Cytolysosome |
| CqLISCL07 | CQ041201 | Cq7A | 662 | 74 879.34 | 6.61 | 细胞核Nuclear |
| CqLISCL08 | CQ032603 | Cq9B | 667 | 75 299.73 | 5.23 | 细胞核Nuclear |
| CqLISCL09 | CQ041200 | Cq7A | 686 | 77 311.71 | 4.87 | 细胞核Nuclear |
| CqLISCL10 | CQ030634 | Cq5A | 760 | 86 162.35 | 5.50 | 细胞核Nuclear |
| CqLISCL11 | CQ055311 | Cq2A | 780 | 87 684.22 | 7.07 | 溶酶体Cytolysosome |
| CqLISCL12 | CQ025002 | Cq1B | 818 | 91 640.62 | 7.49 | 细胞核Nuclear |
| CqLISCL13 | CQ032609 | Cq9B | 756 | 84 422.57 | 5.55 | 细胞核Nuclear |
| CqLISCL14 | CQ041197 | Cq7A | 756 | 84 533.61 | 5.81 | 细胞核Nuclear |
| CqPAT1-01 | CQ024284 | Cq1B | 545 | 61 454.56 | 4.73 | 叶绿体Chloroplast |
| CqPAT1-02 | CQ048458 | Cq1A | 426 | 48 153.43 | 5.27 | 溶酶体Cytolysosome |
| CqPAT1-03 | CQ016853 | Cq3B | 639 | 70 122.51 | 5.46 | 细胞核Nuclear |
| CqPAT1-04 | CQ044170 | Cq3A | 767 | 84 245.91 | 6.39 | 细胞核Nuclear |
| CqPAT1-05 | CQ022180 | Cq4B | 635 | 70 854.40 | 8.16 | 叶绿体Chloroplast |
| CqPAT1-06 | CQ037875 | Cq4A | 634 | 70 854.39 | 7.57 | 叶绿体Chloroplast |
| CqPAT1-07 | CQ010139 | Cq8B | 544 | 61 079.89 | 6.33 | 细胞核Nuclear |
| CqPAT1-08 | CQ038026 | Cq8A | 544 | 61 110.88 | 6.51 | 细胞核Nuclear |
| CqPAT1-09 | CQ013243 | Cq2B | 466 | 52 280.60 | 5.82 | 叶绿体Chloroplast |
| CqPAT1-10 | CQ034654 | Cq9B | 541 | 60 293.76 | 6.56 | 细胞核Nuclear |
| CqPAT1-11 | CQ050106 | Cq9A | 588 | 65 884.16 | 6.69 | 叶绿体Chloroplast |
| CqSCL3L01 | CQ019876 | Cq4B | 454 | 50 710.01 | 6.51 | 细胞核Nuclear |
| CqSCL3L02 | CQ035148 | Cq4A | 454 | 50 837.30 | 6.65 | 细胞核Nuclear |
| CqSCL4L01 | CQ003361 | Cq6B | 557 | 60 789.24 | 4.38 | 细胞核Nuclear |
| CqSCL4L02 | CQ028754 | Cq6A | 610 | 67 157.51 | 4.74 | 细胞核Nuclear |
| CqSCR01 | CQ052618 | Cq9A | 872 | 95 159.47 | 6.39 | 细胞核Nuclear |
| CqSCR02 | CQ007129 | Cq7B | 861 | 93 906.00 | 6.40 | 细胞核Nuclear |
| CqSCR03 | CQ014997 | Cq2B | 396 | 43 813.31 | 6.99 | 叶绿体Chloroplast |
| CqSCR04 | CQ054542 | Cq2A | 396 | 43 859.22 | 7.32 | 细胞核Nuclear |
| CqSHR01 | CQ003291 | Cq6B | 475 | 53 457.35 | 5.88 | 线粒体、细胞骨架 Mitochondrion, cytoskeleton |
| CqSHR02 | CQ028690 | Cq6A | 389 | 43 994.11 | 5.05 | 细胞骨架Cytoskeleton |
| CqSHR03 | CQ018831 | Cq3B | 439 | 50 575.84 | 6.38 | 叶绿体Chloroplast |
| CqSHR04 | CQ044974 | Cq3A | 440 | 50 599.98 | 6.73 | 叶绿体Chloroplast |
| CqSHR05 | CQ003315 | Cq6B | 472 | 53 064.48 | 5.94 | 细胞核Nuclear |
| CqSHR06 | CQ028714 | Cq6A | 467 | 52 415.84 | 5.86 | 细胞核Nuclear |
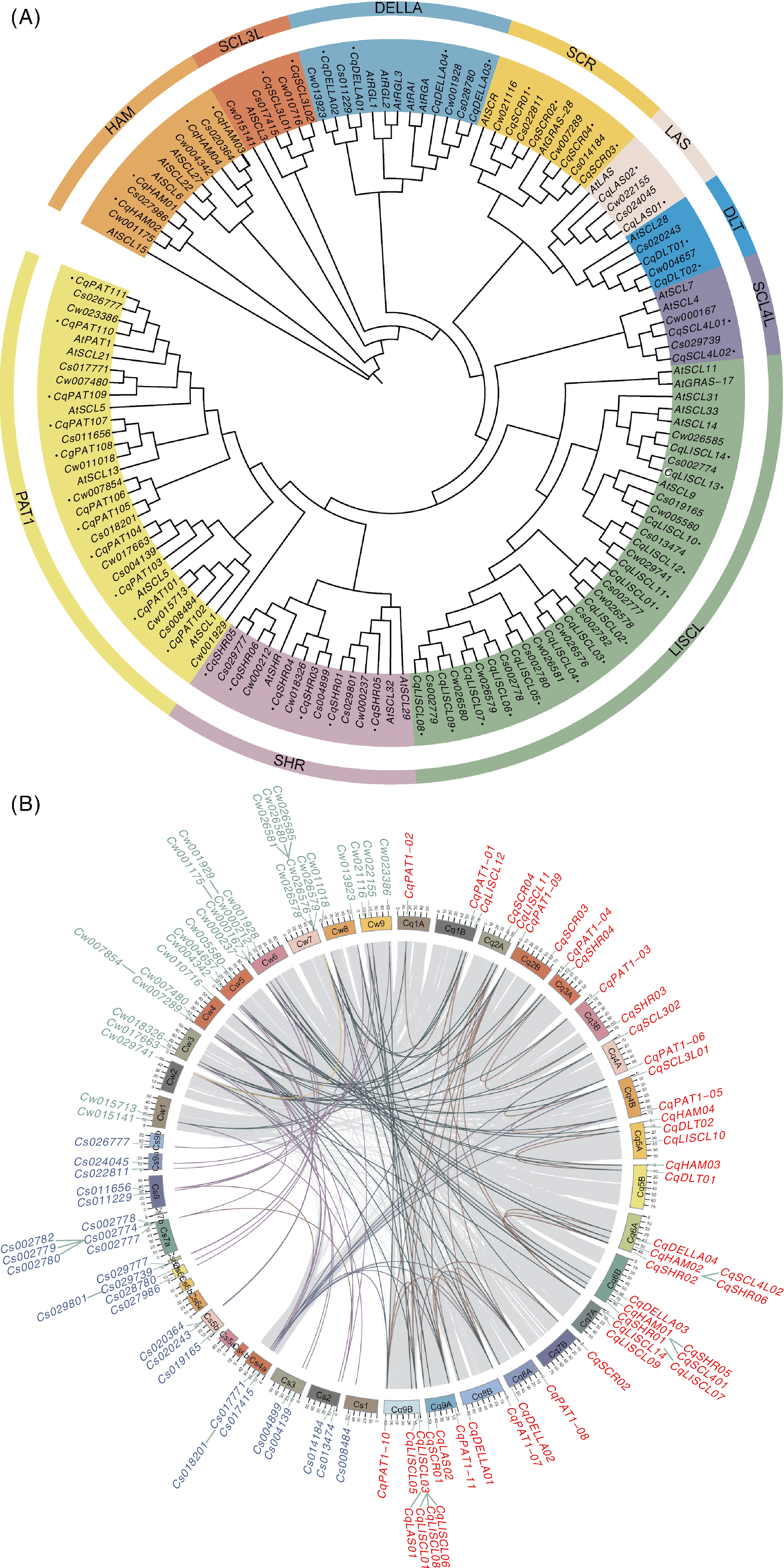
图1 GRAS基因家族系统进化树与共线性分析 A, C. watsonii、C. suecicum、C. quinoa和拟南芥的GRAS家族系统进化树;B,C. watsonii、C. suecicum和C. quinoa的GRAS基因共线性。
Fig.1 Phylogenetic tree and collinearity analysis of GRAS gene family A, Phylogenetic tree of the GRAS family among C. watsonii, C. suecicum, C. quinoa and Arabidopsis thaliana; B, GRAS gene collinearity analysis among C. watsonii, C. suecicum and C. quinoa.
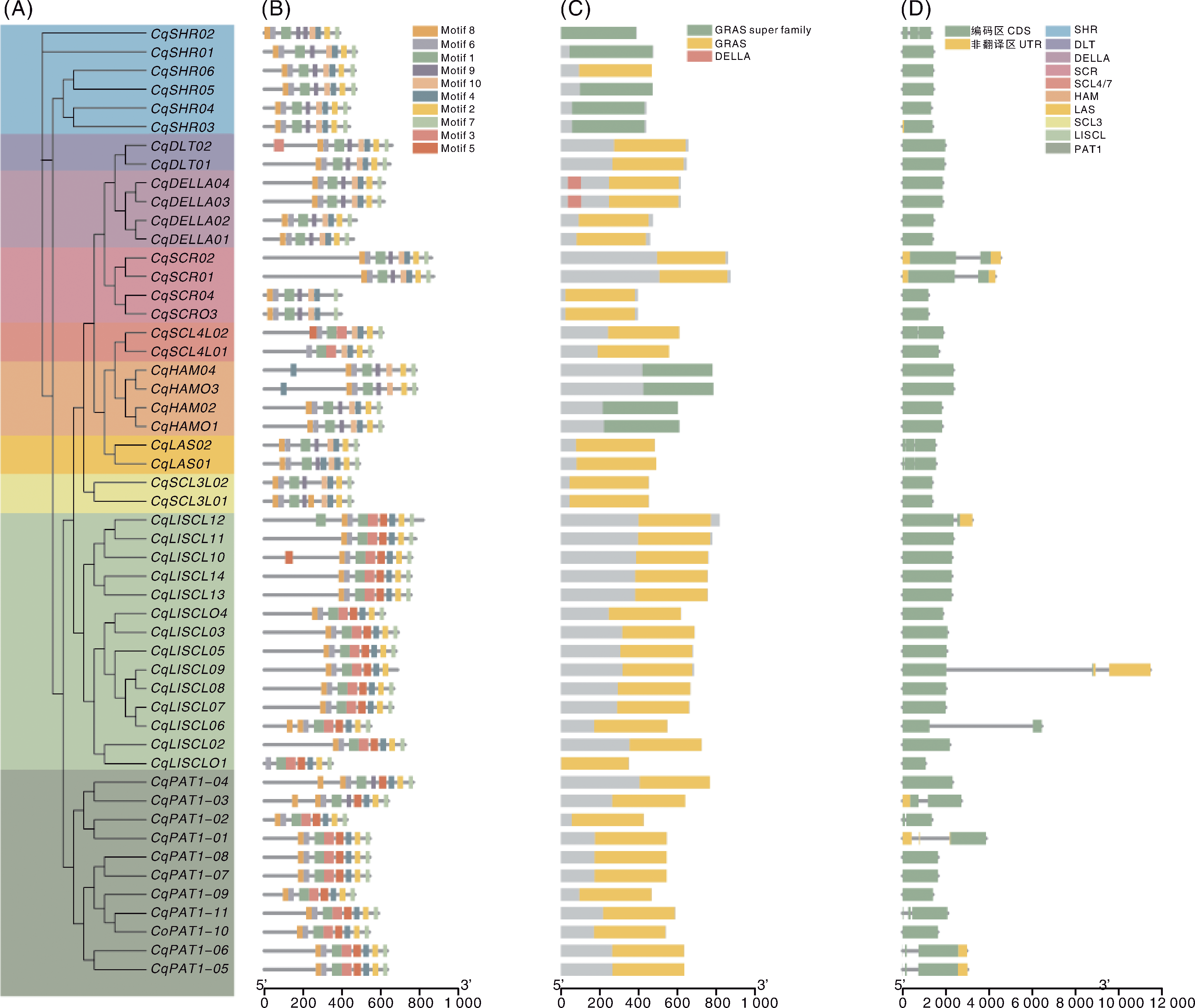
图2 藜麦GRAS基因家族成员聚类、保守基序、保守结构域和基因结构组成分析 A,藜麦GRAS基因家族成员聚类(不同颜色区域代表不同亚家族);B,藜麦GRAS蛋白保守基序组成与分布;C,藜麦GRAS家族蛋白的保守结构域;D,藜麦GRAS基因的结构。
Fig.2 Clustering, conserved motifs, conserved domains, and gene structure composition of quinoa GRAS gene family members A, Clustering of quinoa GRAS gene family members (different colored regions represent different subfamilies); B, Composition and distribution of conserved motifs of CqGRAS; C, Conserved domains of the CqGRAS family proteins; D. Gene structure of CqGRAS.
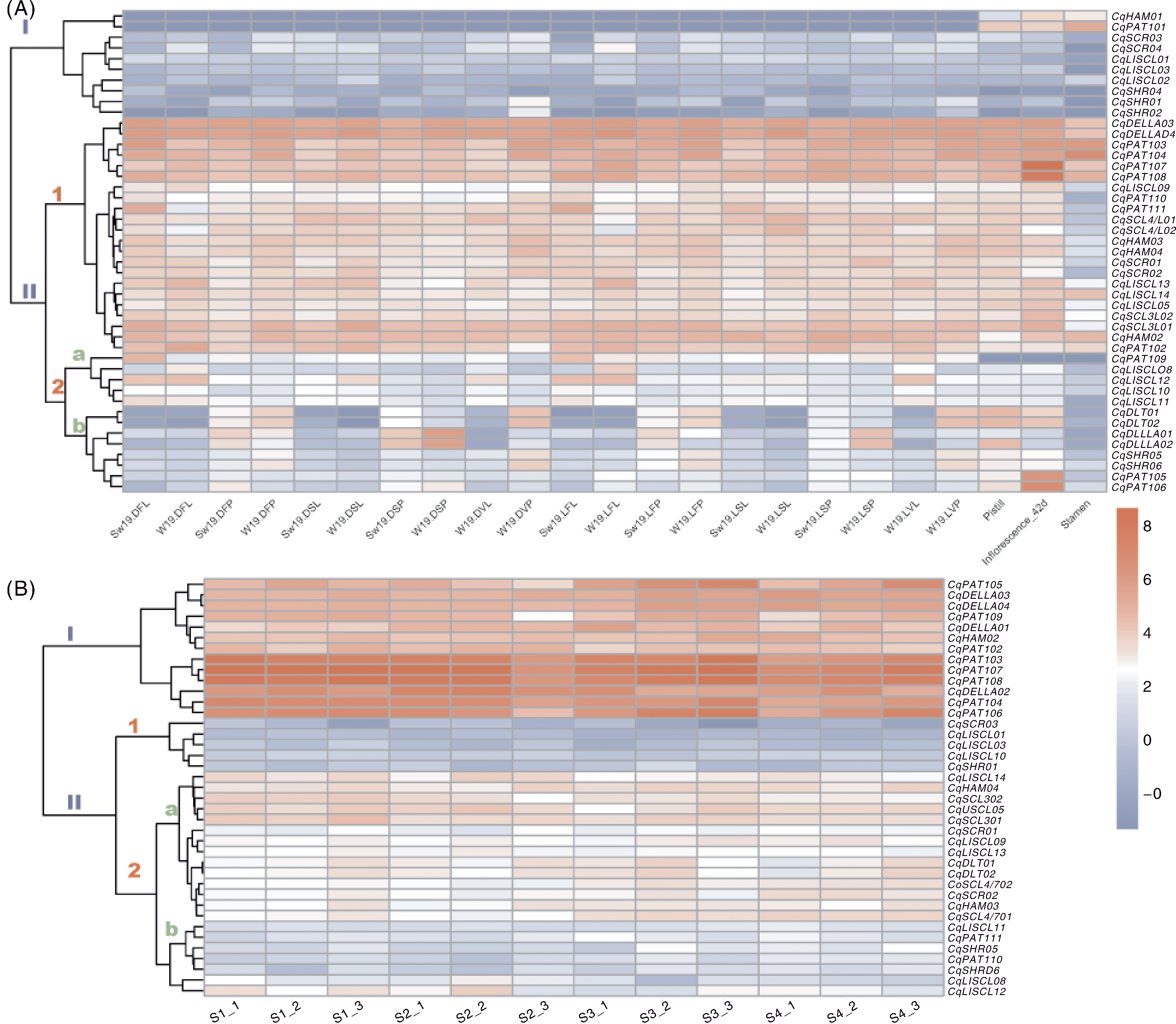
图4 藜麦GRAS基因在不同发育阶段和种子发育过程中的表达 A,不同发育阶段叶片、花序、雄蕊和雌蕊中GRAS基因的表达。样品命名规则:S,夏季;W,冬季;19,W19品种;D,短日(14 h光照/10 h黑暗);L,长日(22 h光照/2 h黑暗);V,营养期;F,开花期;S,种子形成期;P,花序;L,叶片。如S19DFL代表夏季W19短日条件下开花期叶片。B,种子发育阶段GRAS基因的表达。S1~S4代表TY-20开花后7 d(S1)、17 d(S2)、27 d(S3)和42 d(S4)的3次生物学重复(1、2、3)。
Fig.4 Expression of GRAS genes in quinoa at different developmental stages and seed development A, Expression of GRAS genes in leaves, inflorescences, stamens, and pistils at different developmental stages. Sample naming: S, Summer; W, Winter; 19, W19 variety; D, Short-day (14 h light/10 h dark) condition; L, Long-day (22 h light/2 h dark) condition; V, Vegetative stage; F, Flowering stage; S, Seed formation stage; P, Panicle sample; L, Leaf sample. e.g., S19DFL represented summer W19 flowering stage leaves under short-day conditions. B, Expression of GRAS genes during seed development. S1-S4 represent the 7 d (S1), 17 d (S2), 27 d (S3), and 42 d (S4) after flowering in the TY-20, with three biological replicates (1, 2, 3).
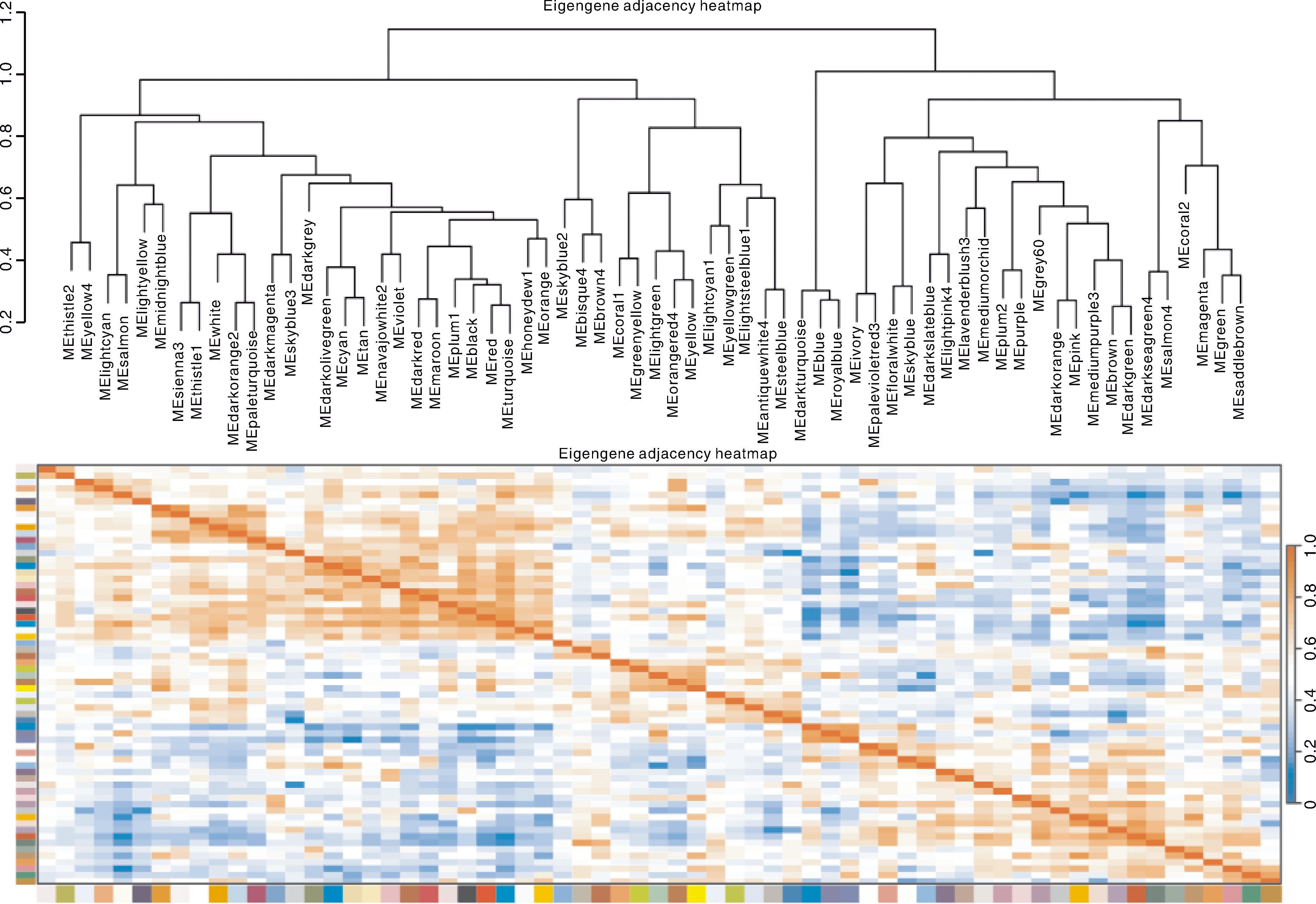
图6 模块与模块之间的聚类和热图 红色的格子代表模块之间具有正相关性,蓝色的格子代表模块之间具有负相关性。
Fig.6 Clustering and heat map between modules Red color of each box represents the positive correlation between modules. Blue color of each box represents the negative correlation between modules.
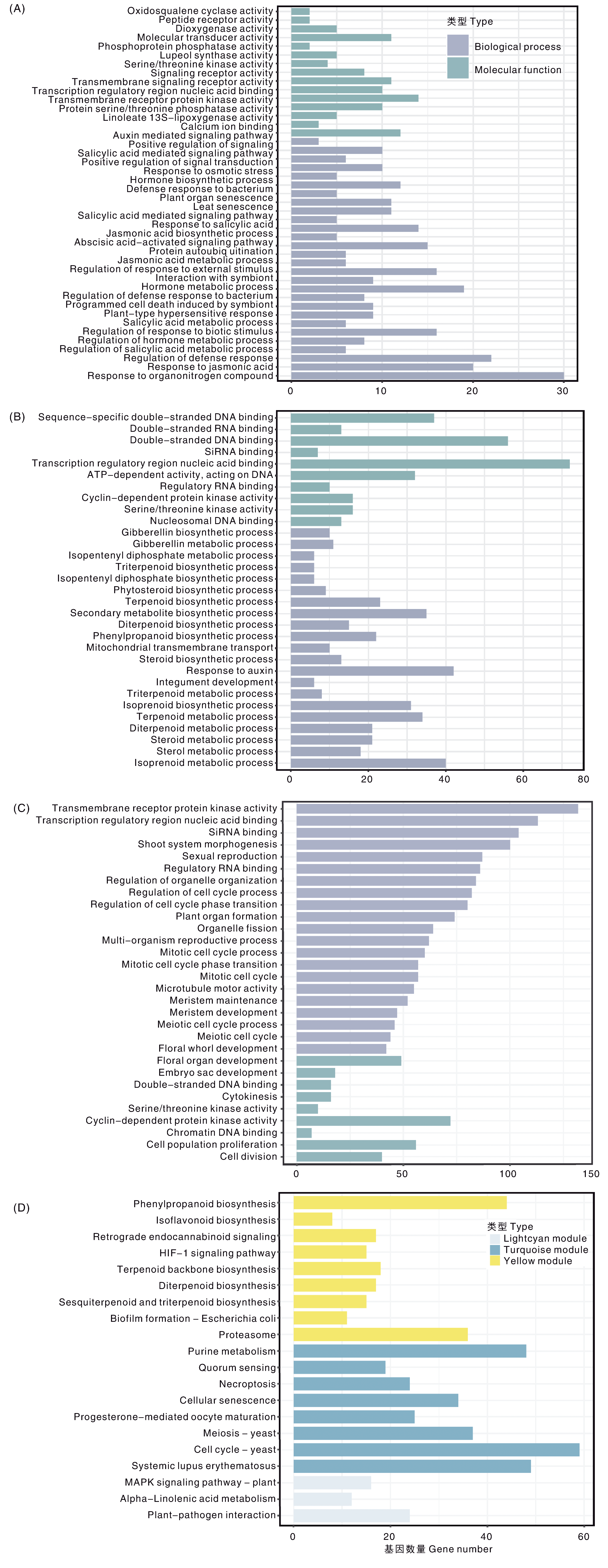
图7 藜麦模块基因GO富集与KEGG富集 A~C分别为lightcyan、yellow和turquoise模块GO富集;D,3个模块共同KEGG富集。
Fig.7 GO enrichment and KEGG enrichment of module genes in quinoa A-C, GO enrichment module of lightcyan, yellow and turquoise, respectively; D, KEGG enrichment of three combining module genes.
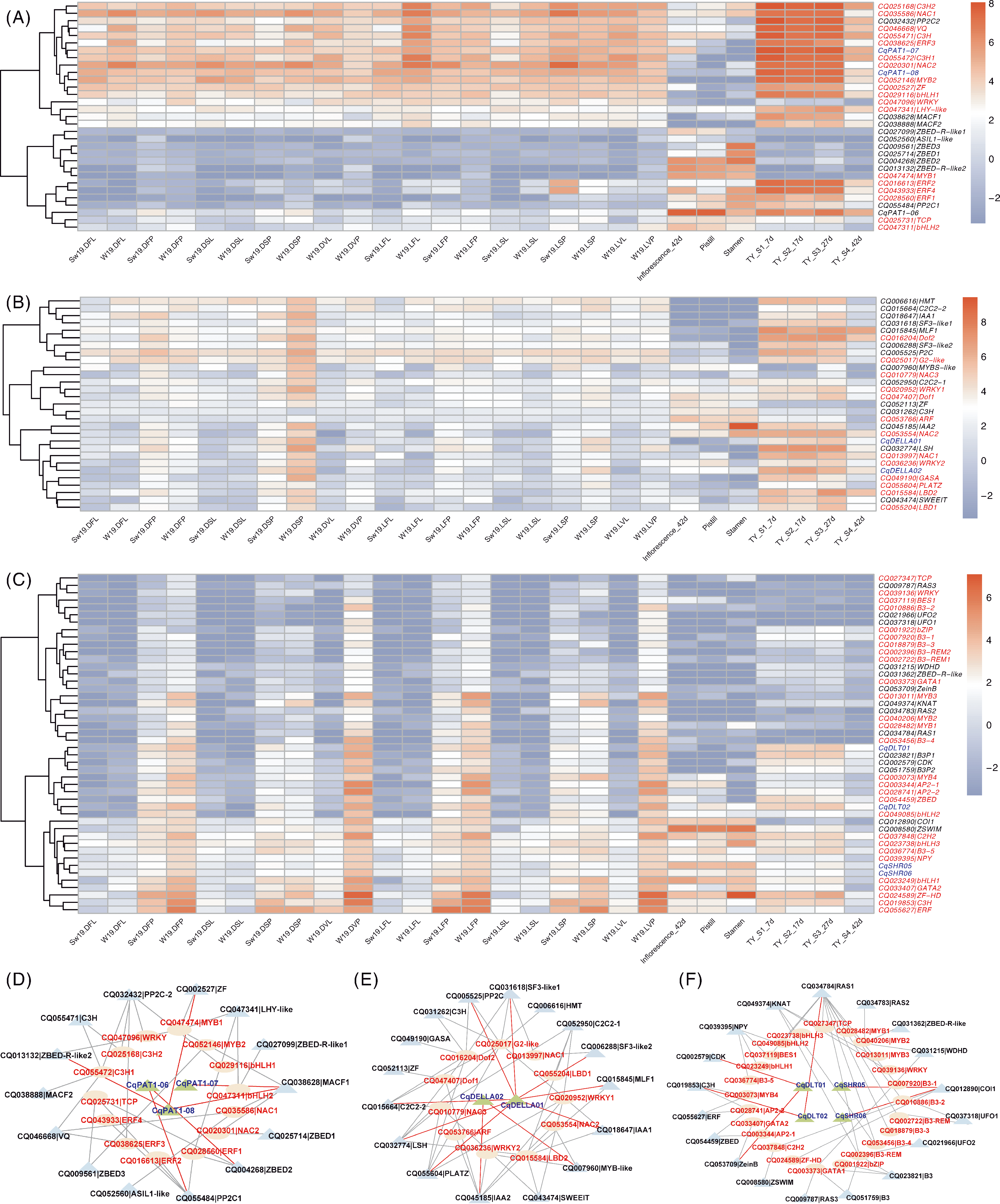
图8 模块中基因与转录因子在花与种子发育相关组织中的表达热图与关系网络 A~C分别代表lightcyan、yellow和tuequoise模块中CqGRAS基因和转录因子在花与种子发育相关组织中的表达;D~F分别代表lightcyan、yellow和tuequoise模块中CqGRAS基因和该模块中相关转录因子的网络图。绿色三角形代表CqGRAS成员,蓝色三角形代表与转录因子互作的其他基因,椭圆形代表转录因子。
Fig.8 Heat map and correlation of gene and transcription factor expression in flower development related tissues within the module A-C represented the expression of CqGRAS genes and transcription factors in flower and seed development related tissues in the lightcyan,yellow and tuequoise modules; D-F represented the network diagram of CqGRAS genes and related transcription factors in the lightcyan,yellow and tuequoise modules. Green triangles represented CqGRAS members, blue triangles represented other genes that interact with transcription factors, and ellipses represented transcription factors.
| [1] | PENG J R, CAROL P, RICHARDS D E, et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses[J]. Genes & Development, 1997, 11(23): 3194-3205. |
| [2] | SILVERSTONE A L, CIAMPAGLIO C N, SUN T P. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway[J]. The Plant Cell, 1998, 10(2): 155-169. |
| [3] | SAMACH A, KLENZ J E, KOHALMI S E, et al. The unusual floral organs gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem[J]. The Plant Journal, 1999, 20(4): 433-445. |
| [4] | BOLLE C. The role of GRAS proteins in plant signal transduction and development[J]. Planta, 2004, 218(5): 683-692. |
| [5] | HIRANO Y, NAKAGAWA M, SUYAMA T, et al. Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD[J]. Nature Plants, 2017, 3: 17010. |
| [6] | PARK M J, KWON Y J, GIL K E, et al. LATE ELONGATED HYPOCOTYL regulates photoperiodic flowering via the circadian clock in Arabidopsis[J]. BMC Plant Biology, 2016, 16(1): 114. |
| [7] | SUN T P, GUBLER F. Molecular mechanism of gibberellin signaling in plants[J]. Annual Review of Plant Biology, 2004, 55: 197-223. |
| [8] | CENCI A, ROUARD M. Evolutionary analyses of GRAS transcription factors in angiosperms[J]. Frontiers in Plant Science, 2017, 8: 273. |
| [9] | MOROHASHI K, MINAMI M, TAKASE H, et al. Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression[J]. Journal of Biological Chemistry, 2003, 278(23): 20865-20873. |
| [10] | SABATINI S, HEIDSTRA R, WILDWATER M, et al. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem[J]. Genes & Development, 2003, 17(3): 354-358. |
| [11] | EDGAR R C. MUSCLE: multiple sequence alignment with high accuracy and high throughput[J]. Nucleic Acids Research, 2004, 32(5): 1792-1797. |
| [12] | TONG H N, JIN Y, LIU W B, et al. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice[J]. The Plant Journal, 2009, 58(5): 803-816. |
| [13] | ZHU X L, WANG B Q, WEI X H. Genome wide identification and expression pattern analysis of the GRAS family in quinoa[J]. Functional Plant Biology, 2021, 48(9): 948-962. |
| [14] | LIU X Y, WIDMER A. Genome-wide comparative analysis of the GRAS gene family in Populus, Arabidopsis and rice[J]. Plant Molecular Biology Reporter, 2014, 32(6): 1129-1145. |
| [15] | LING H, ZENG X, GUO S X. Functional insights into the late embryogenesis abundant (LEA) protein family from Dendrobium officinale(Orchidaceae) using an Escherichia coli system[J]. Scientific Reports, 2016, 6: 39693. |
| [16] | MCGINNIS S, MADDEN T L. BLAST: at the core of a powerful and diverse set of sequence analysis tools[J]. Nucleic Acids Research, 2004, 32: W20-W25. |
| [17] | DUVAUD S, GABELLA C, LISACEK F, et al. Expasy, the Swiss bioinformatics resource portal, as designed by its users[J]. Nucleic Acids Research, 2021, 49(W1): W216-W227. |
| [18] | HORTON P, PARK K J, OBAYASHI T, et al. WoLF PSORT: protein localization predictor[J]. Nucleic Acids Research, 2007, 35: W585-W587. |
| [19] | DREWS G N, BOWMAN J L, MEYEROWITZ E M. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product[J]. Cell, 1991, 65(6): 991-1002. |
| [20] | RUEDA-ROMERO P, BARRERO-SICILIA C, GÓMEZ-CADENAS A, et al. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14[J]. Journal of Experimental Botany, 2012, 63(5): 1937-1949. |
| [21] | URBANOVA T, LEUBNER-METZGER G. Gibberellins and seed germination[M]// HEDDEN P, THOMAS S G. Annual plant reviews. Hoboken: John Wiley & Sons, Inc., 2016: 253-284. |
| [22] | LETUNIC I, BORK P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees[J]. Nucleic Acids Research, 2016, 44(W1): W242-W245. |
| [23] | WANG Y P, TANG H B, DEBARRY J D, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity[J]. Nucleic Acids Research, 2012. 40(7): e49. |
| [24] | CHEN C J, WU Y, XIA R. A painless way to customize Circos plot: from data preparation to visualization using TBtools[J]. iMeta, 2022, 1(3): e35. |
| [25] | HU B, JIN J P, GUO A Y, et al. GSDS 2.0: an upgraded gene feature visualization server[J]. Bioinformatics, 2015, 31(8): 1296-1297. |
| [26] | BAILEY T L, JOHNSON J, GRANT C E, et al. The MEME suite[J]. Nucleic Acids Research, 2015, 43(W1): W39-W49. |
| [27] | MARCHLER-BAUER A, DERBYSHIRE M K, GONZALES N R, et al. CDD: NCBI’s conserved domain database[J]. Nucleic Acids Research, 2015, 43(D1): D222-D226. |
| [28] | CHEN C J, CHEN H, ZHANG Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant, 2020, 13(8): 1194-1202. |
| [29] | SHEN W, SIPOS B, ZHAO L Y. SeqKit2: a Swiss army knife for sequence and alignment processing[J]. iMeta, 2024, 3(3): e191. |
| [30] | LESCOT M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Research, 2002, 30(1): 325-327. |
| [31] | SHAN Z Y, LUO X L, WU M Y, et al. Genome-wide identification and expression of GRAS gene family members in cassava[J]. BMC Plant Biology, 2020, 20(1): 46. |
| [32] | CHEN S F. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp[J]. iMeta, 2023, 2(2): e107. |
| [33] | KIM D, PAGGI J M, PARK C, et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype[J]. Nature Biotechnology, 2019, 37(8): 907-915. |
| [34] | DANECEK P, BONFIELD J K, LIDDLE J, et al. Twelve years of SAMtools and BCFtools[J]. GigaScience, 2021, 10(2): giab008. |
| [35] | LIAO Y, SMYTH G K, SHI W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features[J]. Bioinformatics, 2014, 30(7): 923-930. |
| [36] | DAILEY A L. Metabolomic bioinformatic analysis[M]// Methods in molecular biology. New York: Springer New York, 2017: 341-352. |
| [37] | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔT method[J]. Methods, 2001, 25(4): 402-408. |
| [38] | LANGFELDER P, HORVATH S. WGCNA: an R package for weighted correlation network analysis[J]. BMC Bioinformatics, 2008, 9(1): 559. |
| [39] | WU T Z, HU E Q, XU S B, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data[J]. The Innovation, 2021, 2(3): 100141. |
| [40] | SHANNON P, MARKIEL A, OZIER O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks[J]. Genome Research, 2003, 13(11): 2498-2504. |
| [41] | LEE K C, JANG Y H, KIM S K, et al. RRM domain of Arabidopsis splicing factor SF1 is important for pre-mRNA splicing of a specific set of genes[J]. Plant Cell Reports, 2017, 36(7): 1083-1095. |
| [42] | WILLIAMS J H, DALY L N, INGLEY E, et al. HLS7 a hemopoietic lineage switch gene homologous to the leukemia-inducing gene MLF1[J]. The EMBO Journal, 1999, 18(20): 5559-5566. |
| [43] | CHO E, ZAMBRYSKI P C. ORGAN BOUNDARY1 defines a gene expressed at the junction between the shoot apical meristem and lateral organs[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(5): 2154-2159. |
| [44] | CHEN Y Q, TAI S S, WANG D W, et al. Homology-based analysis of the GRAS gene family in tobacco[J]. Genetics and Molecular Research, 2015, 14(4): 15188-15200. |
| [45] | SONG X M, LIU T K, DUAN W K, et al. Genome-wide analysis of the GRAS gene family in Chinese cabbage (Brassica rapa ssp. pekinensis)[J]. Genomics, 2014, 103(1): 135-146. |
| [46] | STROUD M J, NAZGIEWICZ A, MCKENZIE E A, et al. GAS2-like proteins mediate communication between microtubules and actin through interaction with end-binding proteins[J]. Journal of Cell Science, 2014: jcs.140558. |
| [47] | ZHANG H, LIU X Q, WANG X M, et al. Genome-wide identification of GRAS gene family and their responses to abiotic stress in Medicago sativa[J]. International Journal of Molecular Sciences, 2021, 22(14): 7729. |
| [48] | GAZZARRINI S, LEJAY L, GOJON A, et al. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots[J]. The Plant Cell, 1999, 11(5): 937. |
| [49] | MOURADOV A, CREMER F, COUPLAND G. Control of flowering time: interacting pathways as a basis for diversity[J]. The Plant Cell, 2002, 14(suppl 1): S111-S130. |
| [50] | MOON J, LEE H, KIM M, et al. Analysis of flowering pathway integrators in Arabidopsis[J]. Plant & Cell Physiology, 2005, 46(2): 292-299. |
| [51] | ZHAO H, BAO Y. PIF4: integrator of light and temperature cues in plant growth[J]. Plant Science, 2021, 313: 111086. |
| [52] | BUNDOCK P, HOOYKAAS P. An Arabidopsis hAT-like transposase is essential for plant development[J]. Nature, 2005, 436(7048): 282-284. |
| [53] | CHEN S S, DENG J R, CHENG P D, et al. Transcriptome-wide identification of walnut PP2C family genes in response to external stimulus[J]. BMC Genomics, 2022, 23(1): 640. |
| [54] | DAVIÈRE J M, ACHARD P. Gibberellin signaling in plants[J]. Development, 2013, 140(6): 1147-1151. |
| [55] | MAI Y X, WANG L, YANG H Q. A gain-of-function mutation in IAA7/AXR2 confers late flowering under short-day light in Arabidopsis[J]. Journal of Integrative Plant Biology, 2011, 53(6): 480-492. |
| [56] | RIEU I, RUIZ-RIVERO O, FERNANDEZ-GARCIA N, et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle[J]. The Plant Journal, 2008, 53(3): 488-504. |
| [57] | ZHANG S C, YANG C W, PENG J Z, et al. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana[J]. Plant Molecular Biology, 2009, 69(6): 745-759. |
| [58] | ZHANG Y Q, LIU Z J, LIU J P, et al. GA-DELLA pathway is involved in regulation of nitrogen deficiency-induced anthocyanin accumulation[J]. Plant Cell Reports, 2017, 36(4): 557-569. |
| [59] | MÉNDEZ-BRAVO A, RUIZ-HERRERA L F, CRUZ-RAMÍREZ A, et al. CONSTITUTIVE TRIPLE RESPONSE1 and PIN2 act in a coordinate manner to support the indeterminate root growth and meristem cell proliferating activity in Arabidopsis seedlings[J]. Plant Science, 2019, 280: 175-186. |
| [60] | XU L H, LIU F Q, LECHNER E, et al. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis[J]. The Plant Cell, 2002, 14(8): 1919-1935. |
| [61] | TANG Y X, GAO X, CUI Y N, et al. Research advances in the plant TCP transcription factors[J]. Chinese Science Bulletin, 2022, 67(33): 3964-3975. |
| [62] | KEN H G, TAKANO M, NEUMANN R, et al. The rice COLEOPTILE PHOTOTROPISM1 gene encoding an ortholog of Arabidopsis NPH3 is required for phototropism of coleoptiles and lateral translocation of auxin[J]. The Plant Cell, 2005, 17(1): 103-115. |
| [63] | LEVIN J Z, MEYEROWITZ E M. UFO: an Arabidopsis gene involved in both floral meristem and floral organ development[J]. The Plant Cell, 1995, 7(5): 529. |
| [64] | KROGAN N T, HOGAN K, LONG J A. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19[J]. Development, 2012, 139(22): 4180-4190. |
| [65] | PRESS M O, LANCTOT A, QUEITSCH C. PIF4 and ELF3 act independently in Arabidopsis thaliana thermoresponsive flowering[J]. PLoS One, 2016, 11(8): e0161791. |
| [66] | DALL’OSTO L, CAZZANIGA S, NORTH H, et al. The Arabidopsis Aba4-1 mutant reveals a specific function for neoxanthin in protection against photooxidative stress[J]. The Plant Cell, 2007, 19(3): 1048-1064. |
| [67] | LI X Y, QIAN Q, FU Z M, et al. Control of tillering in rice[J]. Nature, 2003, 422(6932): 618-621. |
| [68] | WANG L, SUN S Y, JIN J Y, et al. Coordinated regulation of vegetative and reproductive branching in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(50): 15504-15509. |
| [69] | VARSHNEY R K, GRANER A, SORRELLS M E. Genomics-assisted breeding for crop improvement[J]. Trends in Plant Science, 2005, 10(12): 621-630. |
| [70] | PEARCE S, SAVILLE R, VAUGHAN S P, et al. Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat[J]. Plant Physiology, 2011, 157(4): 1820-1831. |
| [71] | COLLARD B C Y, MACKILL D J. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century[J]. Philosophical Transactions of the Royal Society B: Biological Sciences, 2008, 363(1491): 557-572. |
| [72] | JARVIS D E, HO Y S, LIGHTFOOT D J, et al. The genome of Chenopodium quinoa[J]. Nature, 2017, 542(7641): 307-312. |
| [73] | RUIZ K B, BIONDI S, MARTÍNEZ E A, et al. Quinoa: a model crop for understanding salt-tolerance mechanisms in halophytes[J]. Plant Biosystems-an International Journal Dealing with All Aspects of Plant Biology, 2016, 150(2): 357-371. |
| [74] | TESTER M, LANGRIDGE P. Breeding technologies to increase crop production in a changing world[J]. Science, 2010, 327(5967): 818-822. |
| [75] | OGATA T, TOYOSHIMA M, YAMAMIZO-ODA C, et al. Virus-mediated transient expression techniques enable functional genomics studies and modulations of betalain biosynthesis and plant height in quinoa[J]. Frontiers in Plant Science, 2021, 12: 643499. |
| [1] | 郑婷, 向江, 魏灵珠, 吴江, 程建徽. 基于WGCNA分析CPPU和TDZ对天工墨玉葡萄香气影响及关键基因挖掘[J]. 浙江农业学报, 2025, 37(2): 311-320. |
| [2] | 周毛措, 卢建雄, 郭晓农, 冯玉兰, 柴薇薇, 高鹏飞. 基于响应面法优化藜麦秸秆发酵工艺[J]. 浙江农业学报, 2024, 36(9): 2020-2030. |
| [3] | 张喜闻, 郭晓农, 王泽兴, 王亚玲. 不同复合益生菌对藜麦秸秆发酵饲料的发酵工艺优化[J]. 浙江农业学报, 2023, 35(12): 2818-2829. |
| [4] | 吕敬, 吴治勇, 郭晓农, 冯玉兰, 卢建雄, 柴薇薇. 基于响应面法的乳酸菌发酵藜麦秸秆工艺条件优化[J]. 浙江农业学报, 2022, 34(9): 1866-1876. |
| [5] | 吴涛, 江小帆, 杨发荣, 魏玉明, 陈国顺, 蔡原, 焦婷, 黄杰, 赵生国. 日粮中不同藜麦添加水平对芦花鸡肉品质及微量元素的影响[J]. 浙江农业学报, 2022, 34(5): 897-907. |
| [6] | 吴涛, 魏玉明, 江小帆, 黄杰, 杨发荣, 陈国顺, 蔡原, 焦婷, 赵生国. 日粮中添加藜麦对芦花鸡生长性能、屠宰性能、器官指数与肠道形态的影响[J]. 浙江农业学报, 2022, 34(2): 255-265. |
| [7] | 时羽杰, 李兴龙, 唐媛, 余海萍, 邬晓勇. 基于GC-MS分析两地白色藜麦种子的代谢差异[J]. 浙江农业学报, 2019, 31(6): 869-877. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||