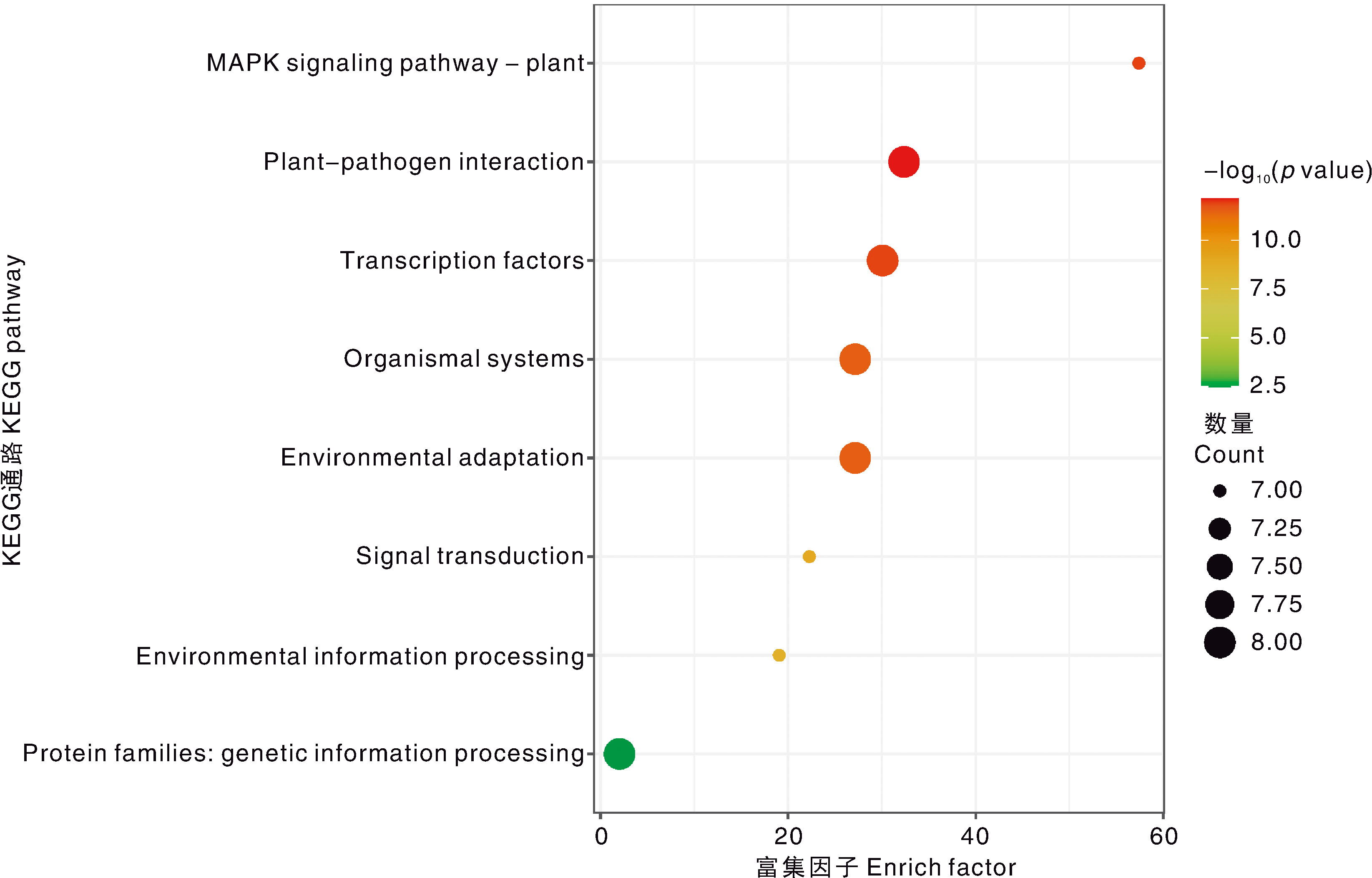
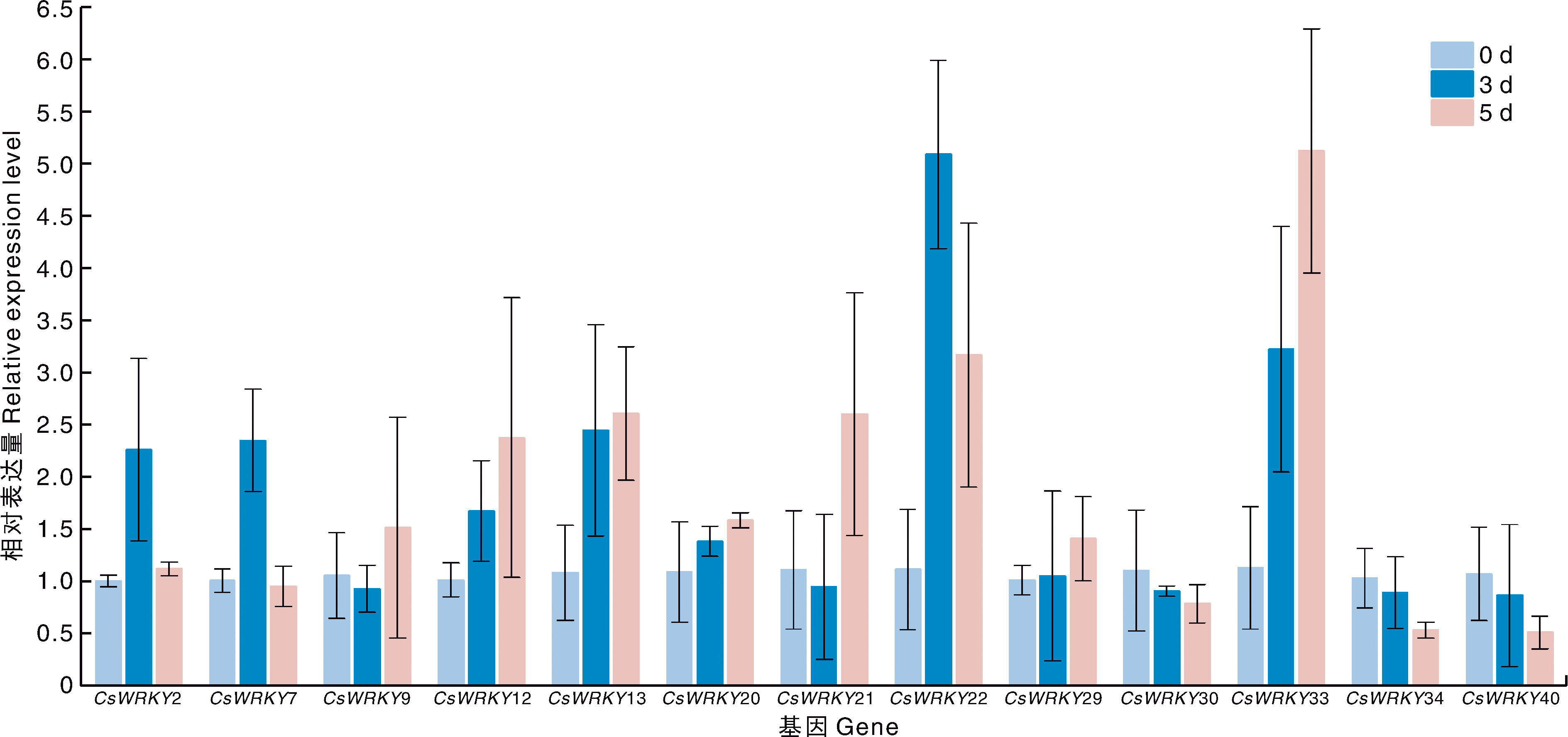
Acta Agriculturae Zhejiangensis ›› 2025, Vol. 37 ›› Issue (10): 2087-2103.DOI: 10.3969/j.issn.1004-1524.20241128
• Horticultural Science • Previous Articles Next Articles
Bioinformatics analysis and drought-tolerant gene mining of WRKY family members in Carex siderosticta
CUI Bowen( ), ZHANG Siyi, WANG Jialing, WANG Jinghong, LIN Jixiang, YANG Qingjie(
), ZHANG Siyi, WANG Jialing, WANG Jinghong, LIN Jixiang, YANG Qingjie( )
)
- College of Landscape Architecture, Northeast Forestry University, Harbin 150040, China
-
Received:2024-12-30Online:2025-10-25Published:2025-11-13
CLC Number:
Cite this article
CUI Bowen, ZHANG Siyi, WANG Jialing, WANG Jinghong, LIN Jixiang, YANG Qingjie. Bioinformatics analysis and drought-tolerant gene mining of WRKY family members in Carex siderosticta[J]. Acta Agriculturae Zhejiangensis, 2025, 37(10): 2087-2103.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.zjnyxb.cn/EN/10.3969/j.issn.1004-1524.20241128
| 基因名称 Gene name | 正向引物序列 Forward primer sequence(5'-3') | 反向引物序列 Reverse primer sequence(5'-3') |
|---|---|---|
| CsEF-1α | AGTGGTTATCGGTCACGTCG | GTTCATCTCAGCGGCTTCCT |
| CsWRKY2 | AATGTGGAGGTTGGTGGGAC | GGACCCGTTGCATCACTCTT |
| CsWRKY7 | GGTTCAAAGAAGCGCGGAAG | AAGTCGGTTTTCGTGCAAGC |
| CsWRKY9 | GCAATCCGAGATGGATCGGT | GCTCTTTTTGCTTCCCGGTG |
| CsWRKY12 | AATGCAGCGGAGTGAAAGGT | TGGTTGTGCTCGCCTTCATA |
| CsWRKY13 | CCAGCTCAGGTTGGTGTTTTG | CCCTCATCCATCCGGCTTAC |
| CsWRKY20 | ACAATGGCTCCAGCAAGACA | ATGATCGGCCTGTGGTTCAG |
| CsWRKY21 | CCACCATGAGGAAAGCTCGT | CTTCAGCACACCTTTGCACC |
| CsWRKY22 | ATGCCTCGTCGTGTTCATCT | CTTGACTTGCTGGAGGTGGT |
| CsWRKY29 | CTAATCACTGCGCCTGACGA | CGTCTTCCCCCGCTTGTAAT |
| CsWRKY30 | GCAGCCACAAAACAAGTCCA | CTGACATTGGTGTCGAGTTGC |
| CsWRKY33 | TTCTTCGTCGTGTGAGGCAA | GTGGCTCACGTTGCCTTTTC |
| CsWRKY34 | ACGGTCAGAAGGTCGTCAAG | GCATGTCGTCCCTCGTATGT |
| CsWRKY40 | GAGGAAGGGTTTTGACGGGT | CCTGGTCGAATTTGTGCGTG |
Table 1 qRT-PCR primer sequences for CsWRKY gene
| 基因名称 Gene name | 正向引物序列 Forward primer sequence(5'-3') | 反向引物序列 Reverse primer sequence(5'-3') |
|---|---|---|
| CsEF-1α | AGTGGTTATCGGTCACGTCG | GTTCATCTCAGCGGCTTCCT |
| CsWRKY2 | AATGTGGAGGTTGGTGGGAC | GGACCCGTTGCATCACTCTT |
| CsWRKY7 | GGTTCAAAGAAGCGCGGAAG | AAGTCGGTTTTCGTGCAAGC |
| CsWRKY9 | GCAATCCGAGATGGATCGGT | GCTCTTTTTGCTTCCCGGTG |
| CsWRKY12 | AATGCAGCGGAGTGAAAGGT | TGGTTGTGCTCGCCTTCATA |
| CsWRKY13 | CCAGCTCAGGTTGGTGTTTTG | CCCTCATCCATCCGGCTTAC |
| CsWRKY20 | ACAATGGCTCCAGCAAGACA | ATGATCGGCCTGTGGTTCAG |
| CsWRKY21 | CCACCATGAGGAAAGCTCGT | CTTCAGCACACCTTTGCACC |
| CsWRKY22 | ATGCCTCGTCGTGTTCATCT | CTTGACTTGCTGGAGGTGGT |
| CsWRKY29 | CTAATCACTGCGCCTGACGA | CGTCTTCCCCCGCTTGTAAT |
| CsWRKY30 | GCAGCCACAAAACAAGTCCA | CTGACATTGGTGTCGAGTTGC |
| CsWRKY33 | TTCTTCGTCGTGTGAGGCAA | GTGGCTCACGTTGCCTTTTC |
| CsWRKY34 | ACGGTCAGAAGGTCGTCAAG | GCATGTCGTCCCTCGTATGT |
| CsWRKY40 | GAGGAAGGGTTTTGACGGGT | CCTGGTCGAATTTGTGCGTG |
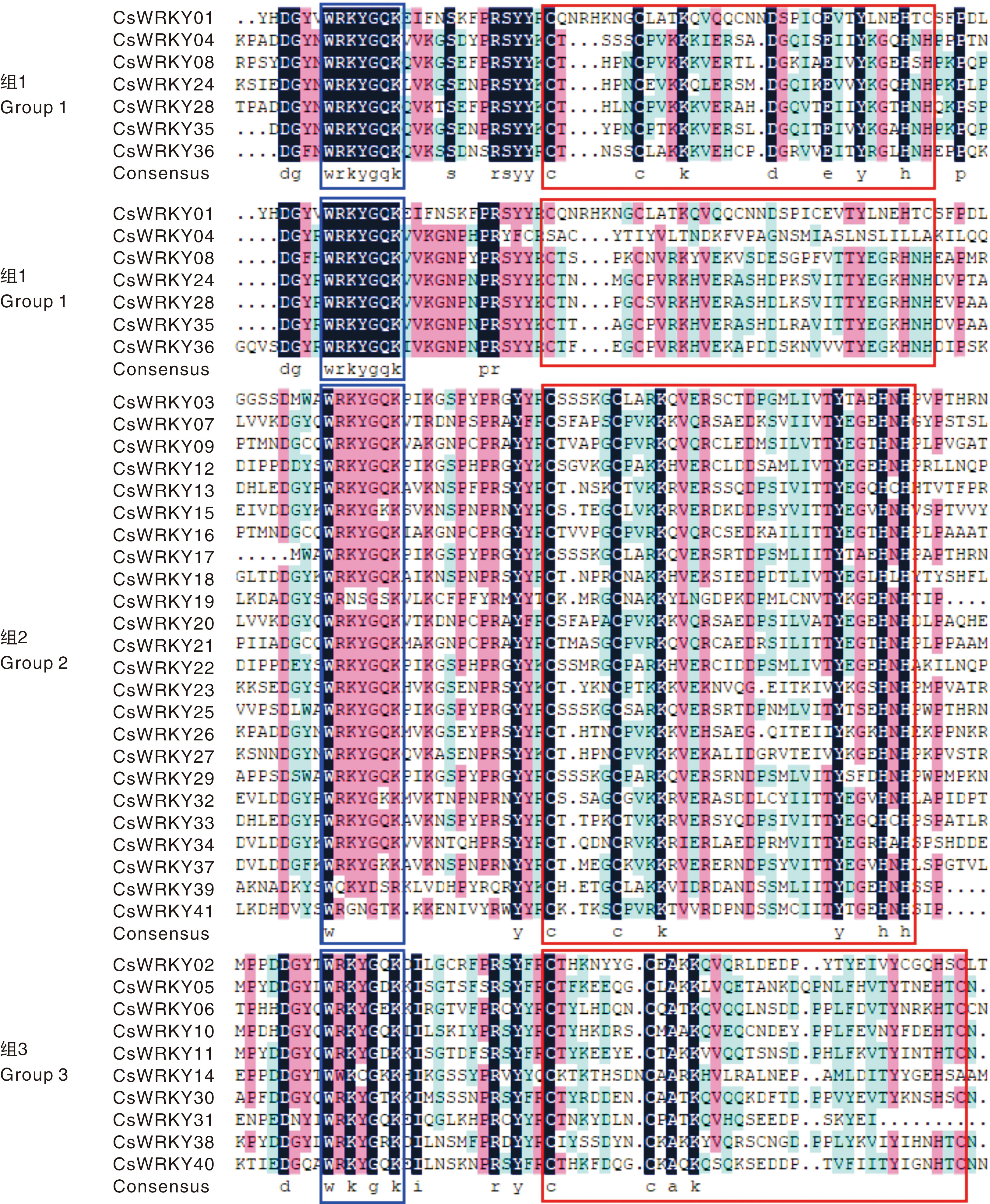
Fig.1 Sequences alignment of conserved domains in the CsWRKY family The WRKY domain is located within the blue box, the zinc finger structure is enclosed within the red box.
| 蛋白质名称 Protein name | 蛋白质ID Protein ID | 氨基酸数量 Number of amino acids | 分子量 Molecular weight/u | 理论等电点 Theoretical pI | 脂肪系数 Aliphatic index | 亲水性指数 Hydropathicity index | 可靠性指数 Reliable index |
|---|---|---|---|---|---|---|---|
| CsWRKY1 | TRINITY_DN25341_c0_g1.p1 | 227 | 25 859.92 | 9.13 | 53.66 | -0.861 | 2.713 |
| CsWRKY2 | TRINITY_DN71036_c0_g1.p1 | 365 | 39 547.20 | 6.26 | 71.32 | -0.387 | 3.220 |
| CsWRKY3 | TRINITY_DN24014_c0_g1.p1 | 345 | 37 300.06 | 5.16 | 72.09 | -0.506 | 4.106 |
| CsWRKY4 | TRINITY_DN21305_c0_g1.p1 | 429 | 46 690.97 | 8.43 | 59.79 | -0.805 | 4.541 |
| CsWRKY5 | TRINITY_DN23710_c0_g1.p1 | 227 | 25 812.35 | 5.98 | 64.89 | -0.558 | 2.769 |
| CsWRKY6 | TRINITY_DN8042_c0_g1.p1 | 1 052 | 114 418.31 | 6.16 | 60.91 | -0.774 | 3.934 |
| CsWRKY7 | TRINITY_DN685_c1_g3.p1 | 320 | 36 371.66 | 7.01 | 63.09 | -0.854 | 4.610 |
| CsWRKY8 | TRINITY_DN13228_c0_g1.p1 | 418 | 46 029.94 | 8.90 | 48.71 | -0.876 | 4.399 |
| CsWRKY9 | TRINITY_DN50400_c0_g1.p1 | 499 | 55 216.80 | 5.22 | 58.10 | -0.941 | 4.819 |
| CsWRKY10 | TRINITY_DN2138_c0_g1.p1 | 287 | 32 783.87 | 6.02 | 72.02 | -0.653 | 4.224 |
| CsWRKY11 | TRINITY_DN35748_c0_g2.p1 | 127 | 15 061.14 | 9.35 | 60.55 | -0.846 | 2.472 |
| CsWRKY12 | TRINITY_DN1223_c0_g2.p1 | 337 | 37 461.27 | 9.54 | 64.51 | -0.785 | 4.270 |
| CsWRKY13 | TRINITY_DN920_c0_g1.p1 | 186 | 20 375.75 | 8.99 | 63.44 | -0.637 | 3.681 |
| CsWRKY14 | TRINITY_DN26245_c0_g2.p1 | 186 | 20 497.80 | 8.25 | 48.82 | -0.712 | 4.341 |
| CsWRKY15 | TRINITY_DN10828_c1_g1.p1 | 236 | 25 423.32 | 5.94 | 54.19 | -0.586 | 3.227 |
| CsWRKY16 | TRINITY_DN52127_c0_g1.p1 | 466 | 51 580.45 | 8.02 | 60.11 | -0.789 | 4.775 |
| CsWRKY17 | TRINITY_DN10234_c0_g1.p1 | 180 | 19 948.94 | 4.83 | 49.39 | -0.923 | 3.832 |
| CsWRKY18 | TRINITY_DN88517_c0_g1.p1 | 222 | 25 105.91 | 6.66 | 61.53 | -0.936 | 4.163 |
| CsWRKY19 | TRINITY_DN60019_c0_g1.p1 | 232 | 26 356.25 | 6.00 | 73.92 | -0.487 | 4.009 |
| CsWRKY20 | TRINITY_DN5707_c0_g1.p1 | 253 | 27 820.48 | 8.97 | 66.68 | -0.733 | 4.411 |
| CsWRKY21 | TRINITY_DN238_c2_g1.p1 | 520 | 55 805.56 | 5.75 | 60.35 | -0.569 | 4.501 |
| CsWRKY22 | TRINITY_DN12424_c0_g1.p1 | 352 | 39 065.32 | 9.84 | 66.22 | -0.782 | 4.599 |
| CsWRKY23 | TRINITY_DN4346_c0_g1.p1 | 309 | 33 839.20 | 7.73 | 52.72 | -0.891 | 4.207 |
| CsWRKY24 | TRINITY_DN8931_c0_g1.p1 | 502 | 54 633.32 | 6.44 | 57.45 | -0.878 | 3.872 |
| CsWRKY25 | TRINITY_DN32124_c3_g1.p1 | 333 | 35 836.41 | 7.66 | 64.23 | -0.535 | 3.024 |
| CsWRKY26 | TRINITY_DN26434_c0_g1.p1 | 352 | 38 775.68 | 6.14 | 50.99 | -0.985 | 4.718 |
| CsWRKY27 | TRINITY_DN714_c2_g1.p1 | 298 | 32 980.16 | 6.20 | 48.12 | -0.912 | 3.346 |
| CsWRKY28 | TRINITY_DN3255_c1_g1.p1 | 722 | 78 422.37 | 5.95 | 48.49 | -0.854 | 3.944 |
| CsWRKY29 | TRINITY_DN4758_c0_g1.p1 | 284 | 30 970.33 | 5.31 | 60.11 | -0.711 | 3.870 |
| CsWRKY30 | TRINITY_DN82013_c0_g1.p1 | 204 | 23 014.52 | 7.04 | 47.79 | -0.749 | 3.481 |
| CsWRKY31 | TRINITY_DN54950_c0_g1.p1 | 121 | 13 931.73 | 9.25 | 62.07 | -1.107 | 2.837 |
| CsWRKY32 | TRINITY_DN44830_c0_g1.p1 | 99 | 11 695.20 | 9.23 | 58.08 | -0.890 | 2.449 |
| CsWRKY33 | TRINITY_DN4854_c0_g1.p1 | 318 | 35 704.86 | 7.11 | 55.50 | -0.876 | 4.841 |
| CsWRKY34 | TRINITY_DN28623_c0_g1.p1 | 234 | 26 682.65 | 9.28 | 51.62 | -0.981 | 4.762 |
| CsWRKY35 | TRINITY_DN15_c2_g1.p1 | 458 | 51 061.95 | 5.85 | 47.53 | -0.967 | 4.421 |
| CsWRKY36 | TRINITY_DN7274_c0_g1.p1 | 394 | 43 242.46 | 8.51 | 57.13 | -0.808 | 4.270 |
| CsWRKY37 | TRINITY_DN9415_c1_g1.p1 | 195 | 22 127.53 | 7.64 | 53.49 | -0.791 | 3.354 |
| CsWRKY38 | TRINITY_DN87366_c0_g1.p1 | 247 | 28 132.69 | 5.20 | 75.75 | -0.517 | 2.689 |
| CsWRKY39 | TRINITY_DN50547_c0_g2.p1 | 115 | 13 177.86 | 9.23 | 58.52 | -0.937 | 2.686 |
| CsWRKY40 | TRINITY_DN5168_c0_g1.p1 | 293 | 32 279.70 | 5.58 | 70.31 | -0.705 | 3.545 |
| CsWRKY41 | TRINITY_DN3189_c2_g1.p1 | 227 | 25 865.76 | 8.86 | 78.06 | -0.437 | 1.947 |
Table 2 Physicochemical properties of CsWRKY proteins
| 蛋白质名称 Protein name | 蛋白质ID Protein ID | 氨基酸数量 Number of amino acids | 分子量 Molecular weight/u | 理论等电点 Theoretical pI | 脂肪系数 Aliphatic index | 亲水性指数 Hydropathicity index | 可靠性指数 Reliable index |
|---|---|---|---|---|---|---|---|
| CsWRKY1 | TRINITY_DN25341_c0_g1.p1 | 227 | 25 859.92 | 9.13 | 53.66 | -0.861 | 2.713 |
| CsWRKY2 | TRINITY_DN71036_c0_g1.p1 | 365 | 39 547.20 | 6.26 | 71.32 | -0.387 | 3.220 |
| CsWRKY3 | TRINITY_DN24014_c0_g1.p1 | 345 | 37 300.06 | 5.16 | 72.09 | -0.506 | 4.106 |
| CsWRKY4 | TRINITY_DN21305_c0_g1.p1 | 429 | 46 690.97 | 8.43 | 59.79 | -0.805 | 4.541 |
| CsWRKY5 | TRINITY_DN23710_c0_g1.p1 | 227 | 25 812.35 | 5.98 | 64.89 | -0.558 | 2.769 |
| CsWRKY6 | TRINITY_DN8042_c0_g1.p1 | 1 052 | 114 418.31 | 6.16 | 60.91 | -0.774 | 3.934 |
| CsWRKY7 | TRINITY_DN685_c1_g3.p1 | 320 | 36 371.66 | 7.01 | 63.09 | -0.854 | 4.610 |
| CsWRKY8 | TRINITY_DN13228_c0_g1.p1 | 418 | 46 029.94 | 8.90 | 48.71 | -0.876 | 4.399 |
| CsWRKY9 | TRINITY_DN50400_c0_g1.p1 | 499 | 55 216.80 | 5.22 | 58.10 | -0.941 | 4.819 |
| CsWRKY10 | TRINITY_DN2138_c0_g1.p1 | 287 | 32 783.87 | 6.02 | 72.02 | -0.653 | 4.224 |
| CsWRKY11 | TRINITY_DN35748_c0_g2.p1 | 127 | 15 061.14 | 9.35 | 60.55 | -0.846 | 2.472 |
| CsWRKY12 | TRINITY_DN1223_c0_g2.p1 | 337 | 37 461.27 | 9.54 | 64.51 | -0.785 | 4.270 |
| CsWRKY13 | TRINITY_DN920_c0_g1.p1 | 186 | 20 375.75 | 8.99 | 63.44 | -0.637 | 3.681 |
| CsWRKY14 | TRINITY_DN26245_c0_g2.p1 | 186 | 20 497.80 | 8.25 | 48.82 | -0.712 | 4.341 |
| CsWRKY15 | TRINITY_DN10828_c1_g1.p1 | 236 | 25 423.32 | 5.94 | 54.19 | -0.586 | 3.227 |
| CsWRKY16 | TRINITY_DN52127_c0_g1.p1 | 466 | 51 580.45 | 8.02 | 60.11 | -0.789 | 4.775 |
| CsWRKY17 | TRINITY_DN10234_c0_g1.p1 | 180 | 19 948.94 | 4.83 | 49.39 | -0.923 | 3.832 |
| CsWRKY18 | TRINITY_DN88517_c0_g1.p1 | 222 | 25 105.91 | 6.66 | 61.53 | -0.936 | 4.163 |
| CsWRKY19 | TRINITY_DN60019_c0_g1.p1 | 232 | 26 356.25 | 6.00 | 73.92 | -0.487 | 4.009 |
| CsWRKY20 | TRINITY_DN5707_c0_g1.p1 | 253 | 27 820.48 | 8.97 | 66.68 | -0.733 | 4.411 |
| CsWRKY21 | TRINITY_DN238_c2_g1.p1 | 520 | 55 805.56 | 5.75 | 60.35 | -0.569 | 4.501 |
| CsWRKY22 | TRINITY_DN12424_c0_g1.p1 | 352 | 39 065.32 | 9.84 | 66.22 | -0.782 | 4.599 |
| CsWRKY23 | TRINITY_DN4346_c0_g1.p1 | 309 | 33 839.20 | 7.73 | 52.72 | -0.891 | 4.207 |
| CsWRKY24 | TRINITY_DN8931_c0_g1.p1 | 502 | 54 633.32 | 6.44 | 57.45 | -0.878 | 3.872 |
| CsWRKY25 | TRINITY_DN32124_c3_g1.p1 | 333 | 35 836.41 | 7.66 | 64.23 | -0.535 | 3.024 |
| CsWRKY26 | TRINITY_DN26434_c0_g1.p1 | 352 | 38 775.68 | 6.14 | 50.99 | -0.985 | 4.718 |
| CsWRKY27 | TRINITY_DN714_c2_g1.p1 | 298 | 32 980.16 | 6.20 | 48.12 | -0.912 | 3.346 |
| CsWRKY28 | TRINITY_DN3255_c1_g1.p1 | 722 | 78 422.37 | 5.95 | 48.49 | -0.854 | 3.944 |
| CsWRKY29 | TRINITY_DN4758_c0_g1.p1 | 284 | 30 970.33 | 5.31 | 60.11 | -0.711 | 3.870 |
| CsWRKY30 | TRINITY_DN82013_c0_g1.p1 | 204 | 23 014.52 | 7.04 | 47.79 | -0.749 | 3.481 |
| CsWRKY31 | TRINITY_DN54950_c0_g1.p1 | 121 | 13 931.73 | 9.25 | 62.07 | -1.107 | 2.837 |
| CsWRKY32 | TRINITY_DN44830_c0_g1.p1 | 99 | 11 695.20 | 9.23 | 58.08 | -0.890 | 2.449 |
| CsWRKY33 | TRINITY_DN4854_c0_g1.p1 | 318 | 35 704.86 | 7.11 | 55.50 | -0.876 | 4.841 |
| CsWRKY34 | TRINITY_DN28623_c0_g1.p1 | 234 | 26 682.65 | 9.28 | 51.62 | -0.981 | 4.762 |
| CsWRKY35 | TRINITY_DN15_c2_g1.p1 | 458 | 51 061.95 | 5.85 | 47.53 | -0.967 | 4.421 |
| CsWRKY36 | TRINITY_DN7274_c0_g1.p1 | 394 | 43 242.46 | 8.51 | 57.13 | -0.808 | 4.270 |
| CsWRKY37 | TRINITY_DN9415_c1_g1.p1 | 195 | 22 127.53 | 7.64 | 53.49 | -0.791 | 3.354 |
| CsWRKY38 | TRINITY_DN87366_c0_g1.p1 | 247 | 28 132.69 | 5.20 | 75.75 | -0.517 | 2.689 |
| CsWRKY39 | TRINITY_DN50547_c0_g2.p1 | 115 | 13 177.86 | 9.23 | 58.52 | -0.937 | 2.686 |
| CsWRKY40 | TRINITY_DN5168_c0_g1.p1 | 293 | 32 279.70 | 5.58 | 70.31 | -0.705 | 3.545 |
| CsWRKY41 | TRINITY_DN3189_c2_g1.p1 | 227 | 25 865.76 | 8.86 | 78.06 | -0.437 | 1.947 |
| 蛋白质名称 Protein name | α螺旋 Alpha-helix | 3_10螺旋 3_10 helix | 延伸链 Extended strand | 氢键转角 Turn | 弯曲 Bend | 无规卷曲 Random coil | β折叠 Beta-sheet | π-螺旋 π-helix |
|---|---|---|---|---|---|---|---|---|
| CsWRKY1 | — | — | 27.31 | 8.81 | 6.17 | 57.71 | — | — |
| CsWRKY2 | 23.84 | — | 10.41 | 2.74 | 2.74 | 60.27 | — | — |
| CsWRKY3 | 15.36 | — | 13.62 | 4.35 | 3.19 | 63.48 | — | — |
| CsWRKY4 | 12.12 | 0.23 | 15.15 | 5.13 | 4.20 | 63.17 | — | — |
| CsWRKY5 | 31.28 | — | 15.42 | 3.52 | 3.52 | 46.26 | — | — |
| CsWRKY6 | 27.76 | 0.38 | 5.99 | 4.09 | 1.33 | 60.46 | — | — |
| CsWRKY7 | 35.94 | 0.94 | 11.25 | 3.75 | 3.44 | 44.69 | — | — |
| CsWRKY8 | 3.11 | — | 17.46 | 3.11 | 5.02 | 71.05 | 0.24 | — |
| CsWRKY9 | 32.67 | — | 9.82 | 2.00 | 2.20 | 53.31 | — | — |
| CsWRKY10 | 27.87 | 1.05 | 10.10 | 4.18 | 3.48 | 53.31 | — | — |
| CsWRKY11 | 5.51 | — | 24.41 | 6.30 | 9.45 | 54.33 | — | — |
| CsWRKY12 | 13.95 | — | 9.20 | 3.56 | 4.15 | 69.14 | — | — |
| CsWRKY13 | 4.84 | 1.61 | 17.74 | 5.91 | 6.45 | 63.44 | — | — |
| CsWRKY14 | 11.29 | 2.15 | 16.13 | 3.76 | 5.38 | 61.29 | — | — |
| CsWRKY15 | 10.59 | 0.42 | 19.07 | 5.08 | 3.39 | 61.44 | — | — |
| CsWRKY16 | 25.11 | — | 8.15 | 2.58 | 3.00 | 61.16 | — | — |
| CsWRKY17 | 3.89 | — | 14.44 | 4.44 | 6.11 | 71.11 | — | — |
| CsWRKY18 | 15.77 | — | 18.47 | 3.15 | 4.50 | 58.11 | — | — |
| CsWRKY19 | 17.67 | — | 15.95 | 5.17 | 5.17 | 56.03 | — | — |
| CsWRKY20 | 18.58 | 1.19 | 15.02 | 3.95 | 3.56 | 57.71 | — | — |
| CsWRKY21 | 18.27 | 0.38 | 8.08 | 4.23 | 3.27 | 65.77 | — | — |
| CsWRKY22 | 24.43 | — | 11.36 | 3.98 | 4.26 | 55.97 | — | — |
| CsWRKY23 | 4.53 | 0.65 | 8.74 | 8.09 | 3.56 | 74.43 | — | — |
| CsWRKY24 | 5.98 | — | 16.53 | 2.99 | 3.78 | 70.72 | — | — |
| CsWRKY25 | 15.92 | 1.50 | 12.91 | 6.91 | 2.40 | 60.36 | — | — |
| CsWRKY26 | 5.40 | 0.57 | 13.35 | 3.98 | 4.26 | 72.16 | 0.28 | — |
| CsWRKY27 | 10.07 | — | 12.42 | 6.04 | 2.68 | 68.79 | — | — |
| CsWRKY28 | 9.00 | 0.28 | 9.56 | 4.43 | 3.19 | 73.55 | — | — |
| CsWRKY29 | 7.39 | 0.70 | 10.92 | 5.99 | 3.52 | 71.48 | — | — |
| CsWRKY30 | 7.35 | — | 17.16 | 4.41 | 3.43 | 67.65 | — | — |
| CsWRKY31 | 18.18 | — | 21.49 | 8.26 | 5.79 | 46.28 | — | — |
| CsWRKY32 | 11.11 | — | 33.33 | 7.07 | 14.14 | 34.34 | — | — |
| CsWRKY33 | 14.15 | 0.31 | 14.47 | 2.83 | 3.14 | 65.09 | — | — |
| CsWRKY34 | 4.27 | — | 14.10 | 2.56 | 5.98 | 73.08 | — | — |
| CsWRKY35 | 6.33 | — | 17.03 | 4.80 | 4.37 | 67.47 | — | — |
| CsWRKY36 | 5.33 | — | 18.53 | 4.06 | 4.82 | 67.26 | — | — |
| CsWRKY37 | 3.08 | — | 23.59 | 3.59 | 5.64 | 64.10 | — | — |
| CsWRKY38 | 23.89 | — | 14.57 | 4.86 | 4.45 | 52.23 | — | — |
| CsWRKY39 | 10.43 | — | 26.96 | 3.48 | 7.83 | 51.30 | — | — |
| CsWRKY40 | 23.89 | — | 10.92 | 5.12 | 2.73 | 57.34 | — | — |
| CsWRKY41 | 19.38 | — | 11.45 | 6.61 | 2.64 | 59.91 | — | — |
Table 3 Proportion of secondary structure of CsWRKY family proteins %
| 蛋白质名称 Protein name | α螺旋 Alpha-helix | 3_10螺旋 3_10 helix | 延伸链 Extended strand | 氢键转角 Turn | 弯曲 Bend | 无规卷曲 Random coil | β折叠 Beta-sheet | π-螺旋 π-helix |
|---|---|---|---|---|---|---|---|---|
| CsWRKY1 | — | — | 27.31 | 8.81 | 6.17 | 57.71 | — | — |
| CsWRKY2 | 23.84 | — | 10.41 | 2.74 | 2.74 | 60.27 | — | — |
| CsWRKY3 | 15.36 | — | 13.62 | 4.35 | 3.19 | 63.48 | — | — |
| CsWRKY4 | 12.12 | 0.23 | 15.15 | 5.13 | 4.20 | 63.17 | — | — |
| CsWRKY5 | 31.28 | — | 15.42 | 3.52 | 3.52 | 46.26 | — | — |
| CsWRKY6 | 27.76 | 0.38 | 5.99 | 4.09 | 1.33 | 60.46 | — | — |
| CsWRKY7 | 35.94 | 0.94 | 11.25 | 3.75 | 3.44 | 44.69 | — | — |
| CsWRKY8 | 3.11 | — | 17.46 | 3.11 | 5.02 | 71.05 | 0.24 | — |
| CsWRKY9 | 32.67 | — | 9.82 | 2.00 | 2.20 | 53.31 | — | — |
| CsWRKY10 | 27.87 | 1.05 | 10.10 | 4.18 | 3.48 | 53.31 | — | — |
| CsWRKY11 | 5.51 | — | 24.41 | 6.30 | 9.45 | 54.33 | — | — |
| CsWRKY12 | 13.95 | — | 9.20 | 3.56 | 4.15 | 69.14 | — | — |
| CsWRKY13 | 4.84 | 1.61 | 17.74 | 5.91 | 6.45 | 63.44 | — | — |
| CsWRKY14 | 11.29 | 2.15 | 16.13 | 3.76 | 5.38 | 61.29 | — | — |
| CsWRKY15 | 10.59 | 0.42 | 19.07 | 5.08 | 3.39 | 61.44 | — | — |
| CsWRKY16 | 25.11 | — | 8.15 | 2.58 | 3.00 | 61.16 | — | — |
| CsWRKY17 | 3.89 | — | 14.44 | 4.44 | 6.11 | 71.11 | — | — |
| CsWRKY18 | 15.77 | — | 18.47 | 3.15 | 4.50 | 58.11 | — | — |
| CsWRKY19 | 17.67 | — | 15.95 | 5.17 | 5.17 | 56.03 | — | — |
| CsWRKY20 | 18.58 | 1.19 | 15.02 | 3.95 | 3.56 | 57.71 | — | — |
| CsWRKY21 | 18.27 | 0.38 | 8.08 | 4.23 | 3.27 | 65.77 | — | — |
| CsWRKY22 | 24.43 | — | 11.36 | 3.98 | 4.26 | 55.97 | — | — |
| CsWRKY23 | 4.53 | 0.65 | 8.74 | 8.09 | 3.56 | 74.43 | — | — |
| CsWRKY24 | 5.98 | — | 16.53 | 2.99 | 3.78 | 70.72 | — | — |
| CsWRKY25 | 15.92 | 1.50 | 12.91 | 6.91 | 2.40 | 60.36 | — | — |
| CsWRKY26 | 5.40 | 0.57 | 13.35 | 3.98 | 4.26 | 72.16 | 0.28 | — |
| CsWRKY27 | 10.07 | — | 12.42 | 6.04 | 2.68 | 68.79 | — | — |
| CsWRKY28 | 9.00 | 0.28 | 9.56 | 4.43 | 3.19 | 73.55 | — | — |
| CsWRKY29 | 7.39 | 0.70 | 10.92 | 5.99 | 3.52 | 71.48 | — | — |
| CsWRKY30 | 7.35 | — | 17.16 | 4.41 | 3.43 | 67.65 | — | — |
| CsWRKY31 | 18.18 | — | 21.49 | 8.26 | 5.79 | 46.28 | — | — |
| CsWRKY32 | 11.11 | — | 33.33 | 7.07 | 14.14 | 34.34 | — | — |
| CsWRKY33 | 14.15 | 0.31 | 14.47 | 2.83 | 3.14 | 65.09 | — | — |
| CsWRKY34 | 4.27 | — | 14.10 | 2.56 | 5.98 | 73.08 | — | — |
| CsWRKY35 | 6.33 | — | 17.03 | 4.80 | 4.37 | 67.47 | — | — |
| CsWRKY36 | 5.33 | — | 18.53 | 4.06 | 4.82 | 67.26 | — | — |
| CsWRKY37 | 3.08 | — | 23.59 | 3.59 | 5.64 | 64.10 | — | — |
| CsWRKY38 | 23.89 | — | 14.57 | 4.86 | 4.45 | 52.23 | — | — |
| CsWRKY39 | 10.43 | — | 26.96 | 3.48 | 7.83 | 51.30 | — | — |
| CsWRKY40 | 23.89 | — | 10.92 | 5.12 | 2.73 | 57.34 | — | — |
| CsWRKY41 | 19.38 | — | 11.45 | 6.61 | 2.64 | 59.91 | — | — |
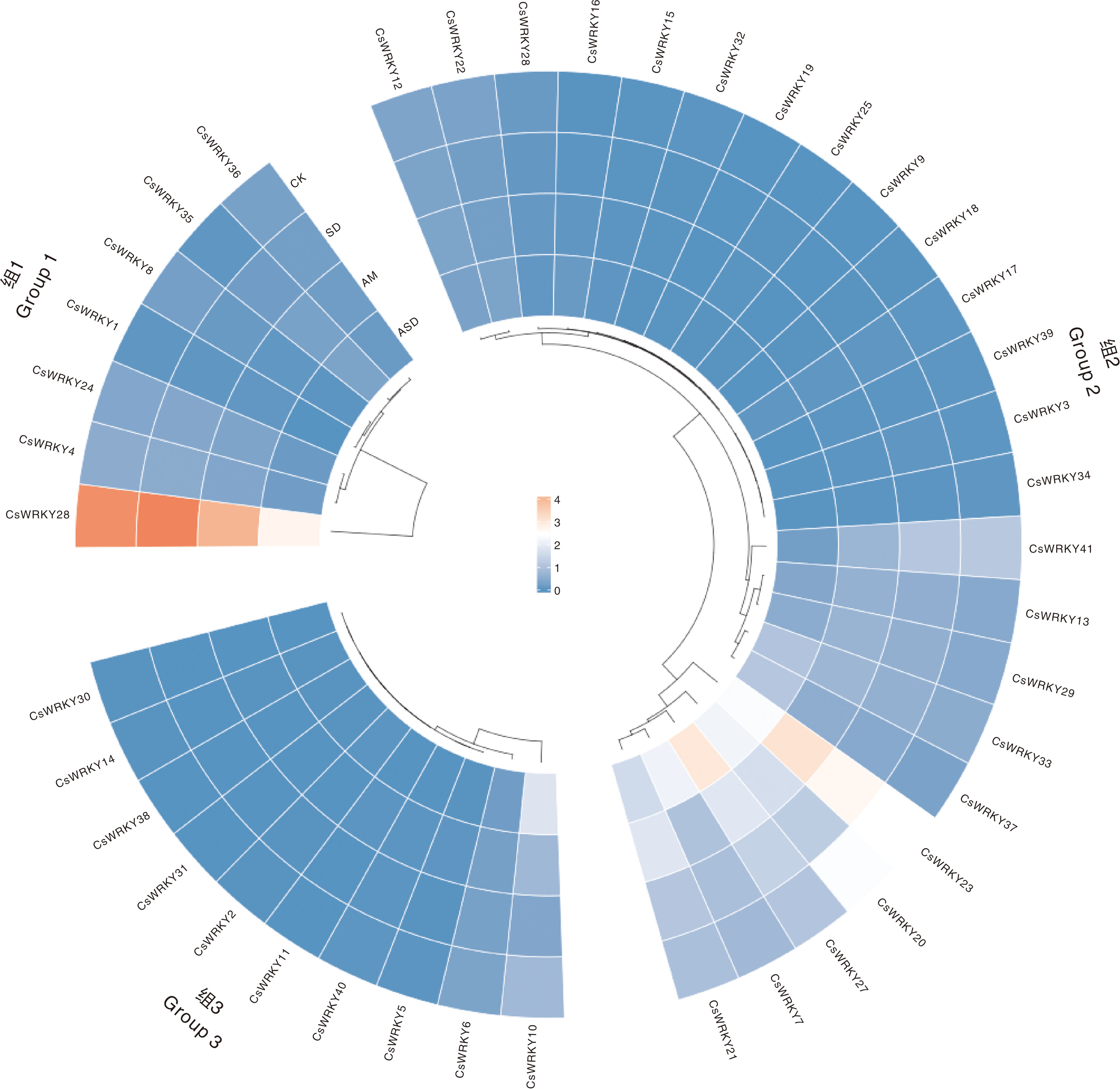
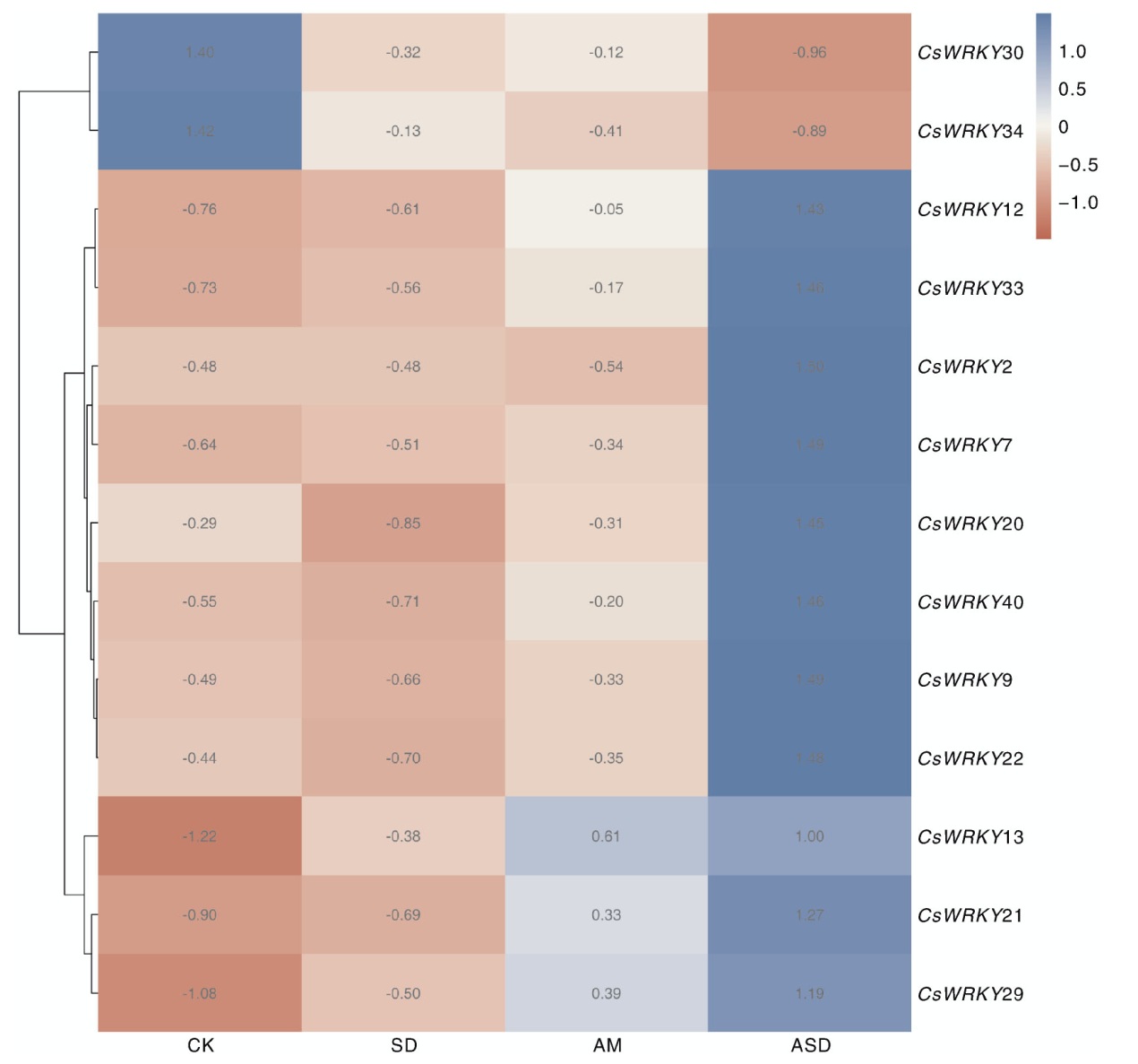
Fig.7 Expression patterns of CsWRKY genes under different treatments CK, Control group; SD, Drought treatment; AM, AM treatment; ASD, AM and drought treatment. The same as in Fig. 8.
| AtWRKY | CsWRKY候选基因 Candidate CsWRKY gene | BLAST位分数 BLAST bit score | AtWRKY | CsWRKY候选基因 Candidate CsWRKY gene | BLAST位分数 BLAST bit score |
|---|---|---|---|---|---|
| AtWRKY11[ | CsWRKY22 CsWRKY12 CsWRKY29 | 186.0 181.0 106.0 | AtWRKY40[ | CsWRKY7 CsWRKY20 CsWRKY9 | 201.0 176.0 103.0 |
| AtWRKY17[ | CsWRKY22 CsWRKY12 CsWRKY29 | 181.0 178.0 103.0 | AtWRKY46[ | CsWRKY40 CsWRKY2 CsWRKY30 | 120.0 92.4 89.0 |
| AtWRKY18[ | CsWRKY7 CsWRKY20 CsWRKY21 | 209.0 171.0 115.0 | AtWRKY57[ | CsWRKY13 CsWRKY33 CsWRKY34 | 154.0 154.0 129.0 |
| AtWRKY28[ | CsWRKY33 CsWRKY13 CsWRKY34 | 182.0 132.0 118.0 | AtWRKY60[ | CsWRKY7 CsWRKY20 CsWRKY9 | 193.0 165.0 107.0 |
| AtWRKY30[ | CsWRKY2 CsWRKY40 CsWRKY30 | 96.7 88.6 81.3 |
Table 4 Candidate CsWRKY genes for drought tolerance
| AtWRKY | CsWRKY候选基因 Candidate CsWRKY gene | BLAST位分数 BLAST bit score | AtWRKY | CsWRKY候选基因 Candidate CsWRKY gene | BLAST位分数 BLAST bit score |
|---|---|---|---|---|---|
| AtWRKY11[ | CsWRKY22 CsWRKY12 CsWRKY29 | 186.0 181.0 106.0 | AtWRKY40[ | CsWRKY7 CsWRKY20 CsWRKY9 | 201.0 176.0 103.0 |
| AtWRKY17[ | CsWRKY22 CsWRKY12 CsWRKY29 | 181.0 178.0 103.0 | AtWRKY46[ | CsWRKY40 CsWRKY2 CsWRKY30 | 120.0 92.4 89.0 |
| AtWRKY18[ | CsWRKY7 CsWRKY20 CsWRKY21 | 209.0 171.0 115.0 | AtWRKY57[ | CsWRKY13 CsWRKY33 CsWRKY34 | 154.0 154.0 129.0 |
| AtWRKY28[ | CsWRKY33 CsWRKY13 CsWRKY34 | 182.0 132.0 118.0 | AtWRKY60[ | CsWRKY7 CsWRKY20 CsWRKY9 | 193.0 165.0 107.0 |
| AtWRKY30[ | CsWRKY2 CsWRKY40 CsWRKY30 | 96.7 88.6 81.3 |
| [1] | HUANG J P, YU H P, DAI A G, et al. Drylands face potential threat under 2 ℃ global warming target[J]. Nature Climate Change, 2017, 7(6): 417-422. |
| [2] | 马住国, 符淙斌, 杨庆, 等. 关于我国北方干旱化及其转折性变化[J]. 大气科学, 2018, 42(4): 951-961. |
| MA Z G, FU C B, YANG Q, et al. Drying trend in Northern China and its shift during 1951-2016[J]. Chinese Journal of Atmospheric Sciences, 2018, 42(4): 951-961. | |
| [3] | 陈兰兰, 王丽, 吴亚娟, 等. 植物响应干旱胁迫的分子和微生态机制[J/OL]. 分子植物育种, 2023: 1-15. [2024-12-10]. https://kns.cnki.net/kcms/detail/46.1068.S.20230406.1634.006.html. |
| CHEN L L, WANG L, WU Y J, et al. Molecular and microecological mechanisms of plant responses to drought stress[J/OL]. Molecular Plant Breeding, 2023: 1-15. [2024-12-10]. https://kns.cnki.net/kcms/detail/46.1068.S.20230406.1634.006.html. (in Chinese with English abstract) | |
| [4] | 刘晨, 徐浩博, 斯钰阳, 等. 基于转录组学的植物响应盐胁迫调控机制研究进展[J]. 浙江农业学报, 2022, 34(4): 870-878. |
| LIU C, XU H B, SI Y Y, et al. Research progress on regulation mechanism of plant response to salt stress based on transcriptomics[J]. Acta Agriculturae Zhejiangensis, 2022, 34(4): 870-878. (in Chinese with English abstract) | |
| [5] | 陈丽飞, 李嘉峻, 刘云怡慧, 等. 干旱胁迫下大苞萱草WRKY基因家族成员鉴定及生物信息学分析[J]. 河南农业科学, 2023, 52(2): 113-123. |
| CHEN L F, LI J J, LIU Y, et al. Identification and bioinformatics analysis of WRKY gene family members in Hemerocallis middendorffii under drought stress[J]. Journal of Henan Agricultural Sciences, 2023, 52(2): 113-123. (in Chinese with English abstract) | |
| [6] | WU K L. The WRKY family of transcription factors in rice and Arabidopsis and their origins[J]. DNA Research, 2005, 12(1): 9-26. |
| [7] | EULGEM T, RUSHTON P J, ROBATZEK S, et al. The WRKY superfamily of plant transcription factors[J]. Trends in Plant Science, 2000, 5(5): 199-206. |
| [8] | BABITHA K C, RAMU S V, PRUTHVI V, et al. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis[J]. Transgenic Research, 2013, 22(2): 327-341. |
| [9] | 张亚萱, 李洪臣, 李丽华, 等. 烟草转录因子NtWRKY65在响应干旱胁迫中的功能研究[J]. 江苏农业科学, 2025, 53(4): 225-233. |
| ZHANG Y X, LI H C, LI L H, et al. Functional analysis of transcription factor NtWRKY65 in tobacco under drought stress[J]. Jiangsu Agricultural Sciences, 2025, 53(4): 225-233. (in Chinese with English abstract) | |
| [10] | 宋娟, 吴祝华, 王靖涵, 等. 丛枝菌根真菌对NaCl胁迫和非胁迫下拟南芥相关指标的影响[J]. 植物资源与环境学报, 2024, 33(6): 113-116. |
| SONG J, WU Z H, WANG J H, et al. Effects of arbuscular mycorrhizal fungi on relative indexes of Arabidopsis thaliana under NaCl stress and non-stress[J]. Journal of Plant Resources and Environment, 2024, 33(6): 113-116. (in Chinese with English abstract) | |
| [11] | QUIROGA G, ERICE G, AROCA R, et al. Elucidating the possible involvement of maize aquaporins and arbuscular mycorrhizal symbiosis in the plant ammonium and urea transport under drought stress conditions[J]. Plants, 2020, 9(2): 148. |
| [12] | SHANG P P, ZHENG R C, LI Y D, et al. Effect of AM fungi on the growth and powdery mildew development of Astragalus sinicus L. under water stress[J]. Plant Physiology and Biochemistry, 2025, 219: 109422. |
| [13] | 钟陈佺. 崖棕抗细菌化学活性成分研究[D]. 杨凌: 西北农林科技大学, 2019. |
| ZHONG C Q. Studies on the anti-bacterial constituents of Carex siderosticta Hance[D]. Yangling: Northwest A & F University, 2019. (in Chinese with English abstract) | |
| [14] | 王艺璇. 冻融循环对宽叶薹草根际土壤与植株生理特性的影响[D]. 哈尔滨: 东北林业大学, 2023. |
| WANG Y X. Effects of freeze-thaw cycles on the physiological characteristics of Carex siderosticta Hance inter-rooted soils and plant[D]. Harbin: Northeast Forestry University, 2023. (in Chinese with English abstract) | |
| [15] | MISTRY J, CHUGURANSKY S, WILLIAMS L, et al. Pfam: the protein families database in 2021[J]. Nucleic Acids Research, 2021, 49(D1): D412-D419. |
| [16] | TAMURA K, PETERSON D, PETERSON N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods[J]. Molecular Biology and Evolution, 2011, 28(10): 2731-2739. |
| [17] | CHEN C J, WU Y, LI J W, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining[J]. Molecular Plant, 2023, 16(11): 1733-1742. |
| [18] | 梁锦, 刘海婷, 钟荣, 等. 萱草不同器官实时荧光定量PCR内参基因的筛选[J]. 植物生理学报, 2020, 56(9): 1891-1898. |
| LIANG J, LIU H T, ZHONG R, et al. Screening of reference genes for quantitative real-time PCR in different organs of Hemerocallis fulva[J]. Plant Physiology Journal, 2020, 56(9): 1891-1898. (in Chinese with English abstract) | |
| [19] | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 - Δ Δ C T method[J]. Methods, 2001, 25(4): 402-408. |
| [20] | ALI M A, AZEEM F, NAWAZ M A, et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis[J]. Journal of Plant Physiology, 2018, 226: 12-21. |
| [21] | CHEN H, LAI Z B, SHI J W, et al. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress[J]. BMC Plant Biology, 2010, 10: 281. |
| [22] | SCARPECI T E, ZANOR M I, MUELLER-ROEBER B, et al. Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana[J]. Plant Molecular Biology, 2013, 83(3): 265-277. |
| [23] | 车永梅, 孙艳君, 卢松冲, 等. AtWRKY40参与拟南芥干旱胁迫响应过程[J]. 植物生理学报, 2018, 54(3): 456-464. |
| CHE Y M, SUN Y J, LU S C, et al. AtWRKY40 functions in drought stress response in Arabidopsis thaliana[J]. Plant Physiology Journal, 2018, 54(3): 456-464. (in Chinese with English abstract) | |
| [24] | DING Z J, YAN J Y, XU X Y, et al. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis[J]. The Plant Journal, 2014, 79(1): 13-27. |
| [25] | JIANG Y J, LIANG G, YU D Q. Activated expression of WRKY57 confers drought tolerance in Arabidopsis[J]. Molecular Plant, 2012, 5(6): 1375-1388. |
| [26] | EULGEM T, SOMSSICH I E. Networks of WRKY transcription factors in defense signaling[J]. Current Opinion in Plant Biology, 2007, 10(4): 366-371. |
| [27] | ROSS C A, LIU Y, SHEN Q J. The WRKY gene family in rice (Oryza sativa)[J]. Journal of Integrative Plant Biology, 2007, 49(6): 827-842. |
| [28] | BENCKE-MALATO M, CABREIRA C, WIEBKE-STROHM B, et al. Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection[J]. BMC Plant Biology, 2014, 14: 236. |
| [29] | 宋辉, 南志标. 蒺藜苜蓿全基因组中WRKY转录因子的鉴定与分析[J]. 遗传, 2014, 36(2): 152-168. |
| SONG H, NAN Z B. Genome-wide identification and analysis of WRKY transcription factors in Medicago truncatula[J]. Hereditas, 2014, 36(2): 152-168. (in Chinese with English abstract) | |
| [30] | WANG H P, CHEN W Q, XU Z Y, et al. Functions of WRKYs in plant growth and development[J]. Trends in Plant Science, 2023, 28(6): 630-645. |
| [31] | ZHOU H, ZHU W, WANG X C, et al. A missense mutation in WRKY32 converts its function from a positive regulator to a repressor of photomorphogenesis[J]. New Phytologist, 2022, 235(1): 111-125. |
| [32] | YIN M, SONG N, CHEN S Y, et al. NaKTI2, a Kunitz trypsin inhibitor transcriptionally regulated by NaWRKY3 and NaWRKY6, is required for herbivore resistance in Nicotiana attenuata[J]. Plant Cell Reports, 2021, 40(1): 97-109. |
| [33] | WANG L J, GUO D Z, ZHAO G D, et al. Group IIc WRKY transcription factors regulate cotton resistance to Fusarium oxysporum by promoting GhMKK2-mediated flavonoid biosynthesis[J]. New Phytologist, 2022, 236(1): 249-265. |
| [34] | LI W X, PANG S Y, LU Z G, et al. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants[J]. Plants, 2020, 9(11): 1515. |
| [35] | FAZAL F M, CHANG H Y. Subcellular spatial transcriptomes: emerging frontier for understanding gene regulation[J]. Cold Spring Harbor Symposia on Quantitative Biology, 2019, 84: 31-45. |
| [36] | 刘晨, 曹小汉, 殷丹丹, 等. MAPK信号通路调控植物响应非生物胁迫的研究进展[J]. 安徽农业科学, 2022, 50(18): 9-16. |
| LIU C, CAO X H, YIN D D, et al. Research progress of MAPK signaling pathway in regulating plants response to abiotic stress[J]. Journal of Anhui Agricultural Sciences, 2022, 50(18): 9-16. (in Chinese with English abstract) | |
| [37] | ZHENG Z Y, MOSHER S L, FAN B F, et al. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae[J]. BMC Plant Biology, 2007, 7: 2. |
| [38] | 王怡, 于月华, 万会娜, 等. GhWRKY44在干旱胁迫下的功能鉴定[J]. 棉花学报, 2024, 36(4): 275-284. |
| WANG Y, YU Y H, WAN H N, et al. Functional identification of GhWRKY44 under drought stress[J]. Cotton Science, 2024, 36(4): 275-284. (in Chinese with English abstract) | |
| [39] | LEE H, CHA J, CHOI C, et al. Rice WRKY11 plays a role in pathogen defense and drought tolerance[J]. Rice, 2018, 11(1): 5. |
| [40] | EL-ESAWI M A, AL-GHAMDI A A, ALI H M, et al. Overexpression of AtWRKY30 transcription factor enhances heat and drought stress tolerance in wheat (Triticum aestivum L.)[J]. Genes, 2019, 10(2): 163. |
| [41] | REN X Z, CHEN Z Z, LIU Y, et al. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis[J]. Plant Journal, 2010, 63(3): 417-429. |
| [42] | QIAO Z, LI C L, ZHANG W. WRKY1 regulates stomatal movement in drought-stressed Arabidopsis thaliana[J]. Plant Molecular Biology, 2016, 91(1): 53-65. |
| [43] | WANG X J, DU B J, LIU M, et al. Arabidopsis transcription factor WRKY33 is involved in drought by directly regulating the expression of CesA8[J]. American Journal of Plant Sciences, 2013, 4(6): 21-27. |
| [1] | LI Yujing, HUANG Qianru, ZHANG Aidong, WU Xuexia, ZHU Dongxing, XIAO Kai. Function of the SmMYB13 gene in drought stress response in eggplant (Solanum melongena L.) [J]. Acta Agriculturae Zhejiangensis, 2025, 37(8): 1666-1679. |
| [2] | SHI Yangyang, LYU Lixia, TUO Dengfeng. Effects of AMF and PGPR on growth and nutrient absorption of Matthiola incana under low temperature and weak light stress [J]. Acta Agriculturae Zhejiangensis, 2025, 37(8): 1694-1705. |
| [3] | DI Yancui, JI Zelin, WANG Yuanyuan, LOU Shihao, ZHANG Tao, GUO Zhixin, SHEN Shunshan, PIAO Fengzhi, DU Nanshan, DONG Xiaoxing, DONG Han. Identification, subcellular localization and expression analysis of tomato SlMYB52 gene [J]. Acta Agriculturae Zhejiangensis, 2025, 37(4): 808-819. |
| [4] | ZHANG Meiying, MO Qian, QI Xiushuang, TONG Ningning, KONG Fan, LIU Zheng’an, LYU Changping, PENG Liping. Cloning and expression analysis of peony PoLPAT2 gene [J]. Acta Agriculturae Zhejiangensis, 2025, 37(2): 321-328. |
| [5] | REN Yuanlong, MA Rong, WANG Xiaozhuo, ZHANG Xueyan. Mitigative effect of foliar spraying melatonin on drought stress of cabbage seedlings [J]. Acta Agriculturae Zhejiangensis, 2025, 37(2): 338-348. |
| [6] | MIN Jiangyan, TANG Zhuolei, YANG Xue, HUANG Xiaoyan, HUANG Kaifeng, HE Peiyun. Effect of different drought-rewatering modes on growth and yield of Tartary buckwheat [J]. Acta Agriculturae Zhejiangensis, 2024, 36(9): 2000-2009. |
| [7] | JIANG Wenjun, SHU Hongsuo, CHEN Zhengman, REN Dianting, YANG Dang, TIAN Rongjiang, DU Zhaokui. Cloning, expression, and bioinformatics analysis of KoWRKY43 gene in Kandelia obovata [J]. Acta Agriculturae Zhejiangensis, 2024, 36(8): 1832-1843. |
| [8] | YANG Mingfeng, JI Chunrong, LIU Yong, BAI Shujun, CHEN Xue, LIU Ailin. Effects of continuous drought stress on cotton growth and soil drought threshold at flowering and boll stage [J]. Acta Agriculturae Zhejiangensis, 2024, 36(4): 738-747. |
| [9] | LI Yaping, JIN Fulai, HUANG Zonggui, ZHANG Tao, DUAN Xiaojing, JIANG Wu, TAO Zhengming, CHEN Jiadong. Identification and expression pattern analysis of glycoside hydrolase GH3 gene family in Dendrobium officinale [J]. Acta Agriculturae Zhejiangensis, 2024, 36(4): 790-799. |
| [10] | ZHANG Luhe, WANG Duofeng, ZHANG De, ZHANG Guangzhong, ZHAO Tong, LYU Binyan, ZHANG Yangjun, LI Yi. Identification and bioinformatics analysis of novel-miR16 target gene ZjTCP4 in Chinese jujube [J]. Acta Agriculturae Zhejiangensis, 2024, 36(3): 534-543. |
| [11] | TIAN Xiaoming, XIANG Guangfeng, MOU Cun, LYU Hao, MA Tao, ZHU Lu, PENG Jing, ZHANG Min, HE Yan. Drought tolerance evaluation of four species of Ormosia [J]. Acta Agriculturae Zhejiangensis, 2024, 36(2): 308-324. |
| [12] | ZHANG Li, WANG Yuanyuan, WANG Rui, LIU Lixia. Cloning sequencing and bioinformatics analysis of DRA gene of yak [J]. Acta Agriculturae Zhejiangensis, 2023, 35(7): 1564-1570. |
| [13] | PANG Xueqing, TANG Shi, ZENG Hongmei, ZHAO Wei, WANG Yin, LUO Yan, YAO Xueping, REN Meishen, REN Yongjun, YANG Zexiao. Cloning and analysis of RdRp gene in two strains of GI.1 and GI.2 RHDV [J]. Acta Agriculturae Zhejiangensis, 2023, 35(6): 1286-1296. |
| [14] | SONG Yaping, LEI Zhaoxiong, ZHAO Yi’ang, JIANG Chao, WANG Xingping, LUORENG Zhuoma, MA Yun, WEI Dawei. Cloning of CDS region of bovine FoxO1 gene and analysing expression pattern during adipocyte differentiation [J]. Acta Agriculturae Zhejiangensis, 2023, 35(5): 1016-1027. |
| [15] | YAO Yanlin, MA Li, LIU Lijun, PU Yuanyuan, LI Xuecai, WANG Wangtian, FANG Yan, SUN Wancang, WU Junyan. Bioinformatics and expression analysis of flowering regulation gene BrFT in Brassica rapa L. [J]. Acta Agriculturae Zhejiangensis, 2023, 35(5): 992-1000. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||