Acta Agriculturae Zhejiangensis ›› 2025, Vol. 37 ›› Issue (12): 2494-2503.DOI: 10.3969/j.issn.1004-1524.20250359
• Animal Science • Previous Articles Next Articles
Molecular characterization and genetic evolutionary analysis of hemagglutinin gene of five H9N2 subtype avian influenza viruses in Zhejiang Province, China
LIU Shizhe1,2( ), HUA Jionggang2, CHEN Liu2, XIE Ronghui3, YE Weicheng2, ZHANG Chuanliang3, ZHU Yinchu2, FENG Xiaoxiao3, FU Yuan2, NI Zheng2, ZHANG Cun2, QU Yonggang1,*(
), HUA Jionggang2, CHEN Liu2, XIE Ronghui3, YE Weicheng2, ZHANG Chuanliang3, ZHU Yinchu2, FENG Xiaoxiao3, FU Yuan2, NI Zheng2, ZHANG Cun2, QU Yonggang1,*( ), YUN Tao2,*(
), YUN Tao2,*( )
)
- 1. College of Animal Science and Technology, Shihezi University, Shihezi 832003, Xinjiang, China
2. Zhejiang Key Laboratory of Livestock and Poultry Biotech Breeding, Key Laboratory of Livestock and Poultry Resources (Poultry) Evaluation and Utilization, Ministry of Agriculture and Rural Affairs, Zhejiang Engineering Research Center for Poultry Breeding Industry and Green Farming Technology, Institute of Animal Husbandry and Veterinary Sciences, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, China
3. Zhejiang Center of Animal Disease Control and Prevention, Hangzhou 311199, China
-
Received:2025-05-06Online:2025-12-25Published:2026-01-09
CLC Number:
Cite this article
LIU Shizhe, HUA Jionggang, CHEN Liu, XIE Ronghui, YE Weicheng, ZHANG Chuanliang, ZHU Yinchu, FENG Xiaoxiao, FU Yuan, NI Zheng, ZHANG Cun, QU Yonggang, YUN Tao. Molecular characterization and genetic evolutionary analysis of hemagglutinin gene of five H9N2 subtype avian influenza viruses in Zhejiang Province, China[J]. Acta Agriculturae Zhejiangensis, 2025, 37(12): 2494-2503.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.zjnyxb.cn/EN/10.3969/j.issn.1004-1524.20250359
| 序号 Number | 参考毒株 Reference strain | 缩写 Abbreviation | HA基因登录号 Accession number |
|---|---|---|---|
| 1 | A/Turkey/Wisconsin/1/1966 | TK/1/66 | CY130054.1 |
| 2 | A/Chicken/Shanghai/F/98 | CK/F/98 | AY743216.1 |
| 3 | A/Chicken/Beijing/1/94 | CK/1/94 | AF156380.1 |
| 4 | A/Chicken/Guangdong/SS/94 | CK/SS/94 | AF384557 |
| 5 | A/Duck/Hong Kong/Y280/97 | DK/Y280/97 | AF156376.1 |
| 6 | A/Turkey/California/189/66 | TK/189/66 | AF156390.1 |
| 7 | A/Shorebird/Delaware/9/96 | SB/9/96 | AF156386.1 |
| 8 | A/Chicken/Korea/SH0802/2008 | CK/SH/08 | HQ221677.1 |
| 9 | A/Chicken/Korea/MS96-CE6/1996 | CK/MS/96 | GU053186.1 |
| 10 | A/Chicken/Israel/178/2006 | CK/178/06 | EF492224.1 |
| 11 | A/Chicken/Karaj/GHA/2017 | CK/GHA/17 | MH360730.1 |
| 12 | A/Chicken/Henan/43/02 | CK/43/02 | DQ064369.1 |
| 13 | A/Goose/China/JY100405/2018 | GS/JY/18 | OQ804402.1 |
| 14 | A/Chicken/Jiangsu/J2303/2018 | CK/J2303/18 | MK553246.1 |
| 15 | A/Chicken/Rizhao/1436/2013 | CK/1436/13 | KF259177.1 |
| 16 | A/Chicken/China/GD2021/2021 | CK/GD2021/21 | MW548848.1 |
| 17 | A/Chicken/Daye/DY0602/2017 | CK/DY0602/17 | MF794999.1 |
| 18 | A/Chicken/China/355/2017 | CK/355/17 | MN385379.1 |
| 19 | A/Chicken/Guangzhou/18/2019 | CK/18/19 | OQ054042.1 |
| 20 | A/Chicken/China/D3/2022 | CK/D3/22 | OR528233.1 |
| 21 | A/Duck/Guangdong/FS91/2022 | DK/FS91/22 | OQ826094.1 |
| 22 | A/Environment/Xiamen/01/2021 | EN/01/21 | ON856628.1 |
| 23 | A/Chicken/Shandong/6/96 | CK/6/96 | DQ064376.1 |
| 24 | A/Chicken/Shandong/1/2000 | CK/1/00 | FJ793404.1 |
| 25 | A/Chicken/Jilin/A/2012 | CK/A/12 | KF886521.1 |
| 26 | A/Chicken/Jiangxi/X1459/2018 | CK/X1459/18 | MK552958.1 |
| 27 | A/Chicken/Guizhou/B11/2011 | CK/B11/11 | JN804200.1 |
| 28 | A/Chicken/Guangdong/WBQ02/2012 | CK/WB/12 | KJ768997.1 |
Table 1 Basic information of reference strain used in this study
| 序号 Number | 参考毒株 Reference strain | 缩写 Abbreviation | HA基因登录号 Accession number |
|---|---|---|---|
| 1 | A/Turkey/Wisconsin/1/1966 | TK/1/66 | CY130054.1 |
| 2 | A/Chicken/Shanghai/F/98 | CK/F/98 | AY743216.1 |
| 3 | A/Chicken/Beijing/1/94 | CK/1/94 | AF156380.1 |
| 4 | A/Chicken/Guangdong/SS/94 | CK/SS/94 | AF384557 |
| 5 | A/Duck/Hong Kong/Y280/97 | DK/Y280/97 | AF156376.1 |
| 6 | A/Turkey/California/189/66 | TK/189/66 | AF156390.1 |
| 7 | A/Shorebird/Delaware/9/96 | SB/9/96 | AF156386.1 |
| 8 | A/Chicken/Korea/SH0802/2008 | CK/SH/08 | HQ221677.1 |
| 9 | A/Chicken/Korea/MS96-CE6/1996 | CK/MS/96 | GU053186.1 |
| 10 | A/Chicken/Israel/178/2006 | CK/178/06 | EF492224.1 |
| 11 | A/Chicken/Karaj/GHA/2017 | CK/GHA/17 | MH360730.1 |
| 12 | A/Chicken/Henan/43/02 | CK/43/02 | DQ064369.1 |
| 13 | A/Goose/China/JY100405/2018 | GS/JY/18 | OQ804402.1 |
| 14 | A/Chicken/Jiangsu/J2303/2018 | CK/J2303/18 | MK553246.1 |
| 15 | A/Chicken/Rizhao/1436/2013 | CK/1436/13 | KF259177.1 |
| 16 | A/Chicken/China/GD2021/2021 | CK/GD2021/21 | MW548848.1 |
| 17 | A/Chicken/Daye/DY0602/2017 | CK/DY0602/17 | MF794999.1 |
| 18 | A/Chicken/China/355/2017 | CK/355/17 | MN385379.1 |
| 19 | A/Chicken/Guangzhou/18/2019 | CK/18/19 | OQ054042.1 |
| 20 | A/Chicken/China/D3/2022 | CK/D3/22 | OR528233.1 |
| 21 | A/Duck/Guangdong/FS91/2022 | DK/FS91/22 | OQ826094.1 |
| 22 | A/Environment/Xiamen/01/2021 | EN/01/21 | ON856628.1 |
| 23 | A/Chicken/Shandong/6/96 | CK/6/96 | DQ064376.1 |
| 24 | A/Chicken/Shandong/1/2000 | CK/1/00 | FJ793404.1 |
| 25 | A/Chicken/Jilin/A/2012 | CK/A/12 | KF886521.1 |
| 26 | A/Chicken/Jiangxi/X1459/2018 | CK/X1459/18 | MK552958.1 |
| 27 | A/Chicken/Guizhou/B11/2011 | CK/B11/11 | JN804200.1 |
| 28 | A/Chicken/Guangdong/WBQ02/2012 | CK/WB/12 | KJ768997.1 |
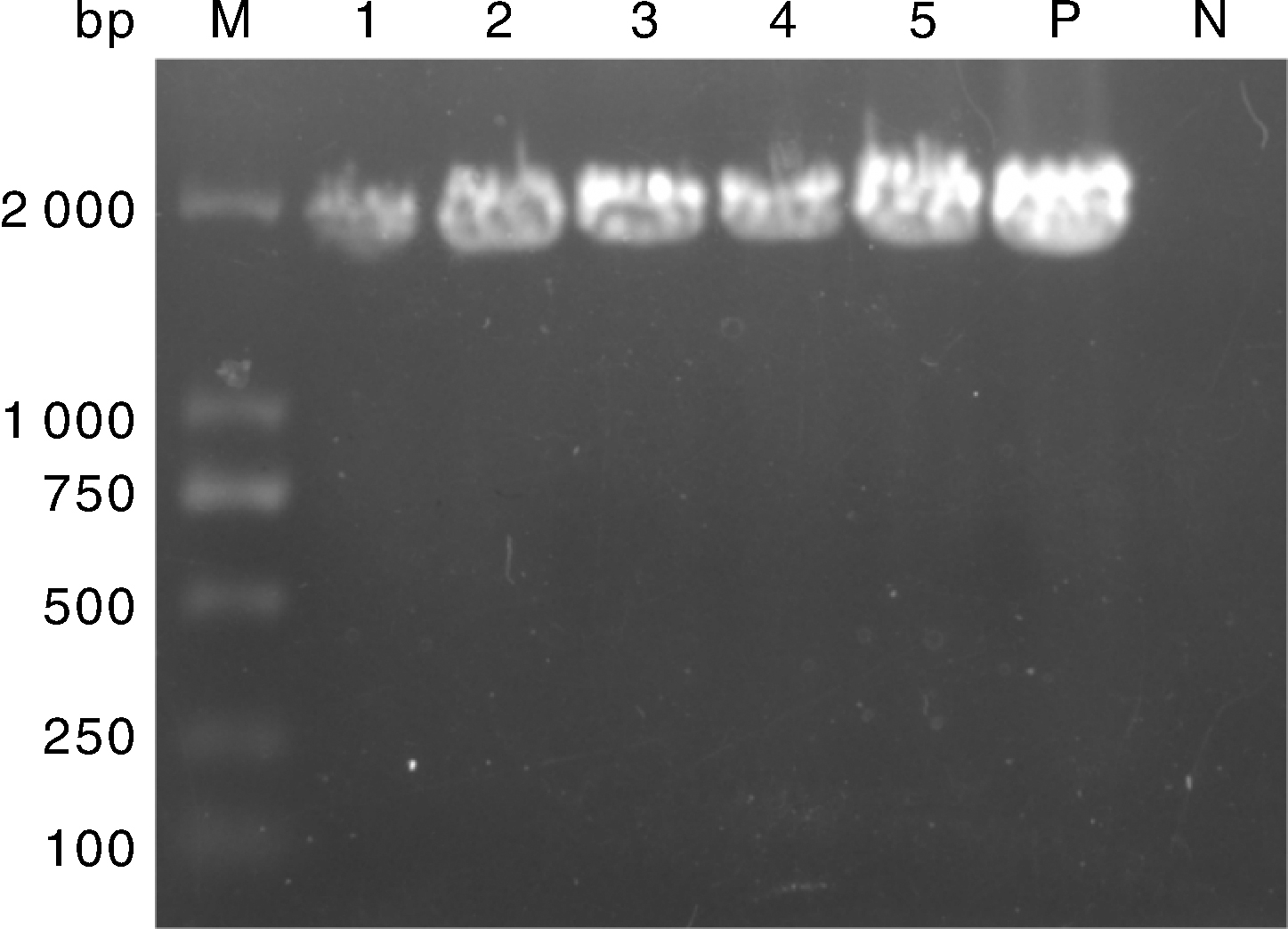
Fig.1 RT-PCR results of HA gene of five H9N2 subtype AIV isolates M, DL2000 DNA marker; 1, CK/01/22 isolate; 2, QU/01/12 isolate; 3, QU/535/12 isolate; 4, CK/541/18 isolate; 5, DK/01/22 isolate; P, Positive control; N, Negative control.
| 毒株 Strain | 分离 时间 Time | 地区 Area | 宿主 Specie | 基因 Gene | 同源性最高的病毒株 Viral strain with the highest homology | 核酸同源性 Nucleotide homology/% | 氨基酸同源性 Amino acid homology/% | 登录号 Accession number |
|---|---|---|---|---|---|---|---|---|
| A/Chicken/Zhejiang/ HZ01/2022 (CK/01/22) | 2022 | 杭州 Hangzhou | 鸡 Gallus demesticus | HA | A/Chicken/Jiangxi/ X1459/2018(H9N2) | 98.75 | 99.29 | MK552958.1 |
| A/Quails/Zhejiang/ SY01/2012 (QU/01/12) | 2012 | 上虞 Shangyu | 鹌鹑 Coturnix japonica | HA | A/Chicken/Shanghai/ S2277/2017(H9N2) | 98.70 | 97.39 | MK554249.1 |
| A/Quails/Zhejiang/ JX535/2012 (QU/535/12) | 2012 | 嘉兴 Jiaxing | 鹌鹑 Coturnix japonica | HA | A/Chicken/Shanghai/ S2277/2017(H9N2) | 98.88 | 98.32 | MK554249.1 |
| A/Chicken/Zhejiang/ XS541/2018 (CK/541/18) | 2018 | 象山 Xiangshan | 鸡 Gallus demesticus | HA | A/Chicken/Jilin/ A/2012(H9N2) | 99.81 | 99.44 | KF886521.1 |
| A/Duck/Zhejiang/ TZ01/2022 (DK/01/22) | 2022 | 台州 Taizhu | 鸭 Anas platyrhynchos | HA | A/Chicken/Jiangxi/ X1459/2018(H9N2) | 99.75 | 99.29 | MK552958.1 |
Table 2 Basic information on the five H9 subtype AIV isolates and their most homologous strains
| 毒株 Strain | 分离 时间 Time | 地区 Area | 宿主 Specie | 基因 Gene | 同源性最高的病毒株 Viral strain with the highest homology | 核酸同源性 Nucleotide homology/% | 氨基酸同源性 Amino acid homology/% | 登录号 Accession number |
|---|---|---|---|---|---|---|---|---|
| A/Chicken/Zhejiang/ HZ01/2022 (CK/01/22) | 2022 | 杭州 Hangzhou | 鸡 Gallus demesticus | HA | A/Chicken/Jiangxi/ X1459/2018(H9N2) | 98.75 | 99.29 | MK552958.1 |
| A/Quails/Zhejiang/ SY01/2012 (QU/01/12) | 2012 | 上虞 Shangyu | 鹌鹑 Coturnix japonica | HA | A/Chicken/Shanghai/ S2277/2017(H9N2) | 98.70 | 97.39 | MK554249.1 |
| A/Quails/Zhejiang/ JX535/2012 (QU/535/12) | 2012 | 嘉兴 Jiaxing | 鹌鹑 Coturnix japonica | HA | A/Chicken/Shanghai/ S2277/2017(H9N2) | 98.88 | 98.32 | MK554249.1 |
| A/Chicken/Zhejiang/ XS541/2018 (CK/541/18) | 2018 | 象山 Xiangshan | 鸡 Gallus demesticus | HA | A/Chicken/Jilin/ A/2012(H9N2) | 99.81 | 99.44 | KF886521.1 |
| A/Duck/Zhejiang/ TZ01/2022 (DK/01/22) | 2022 | 台州 Taizhu | 鸭 Anas platyrhynchos | HA | A/Chicken/Jiangxi/ X1459/2018(H9N2) | 99.75 | 99.29 | MK552958.1 |
| 毒株 Strain | 核苷酸同源性Nucleotide homology | 氨基酸同源性Amino acid homology | ||||||
|---|---|---|---|---|---|---|---|---|
| CK/SS/94 | CK/F/98 | CK/6/96 | CK/1/00 | CK/SS/94 | CK/F/98 | CK/6/96 | CK/1/00 | |
| CK/01/22 | 88.35 | 87.88 | 87.70 | 87.44 | 90.18 | 90.18 | 89.64 | 89.62 |
| QU/01/12 | 87.84 | 86.60 | 86.78 | 86.78 | 89.70 | 88.39 | 88.80 | 88.53 |
| QU/535/12 | 88.46 | 87.21 | 88.08 | 87.41 | 90.82 | 89.14 | 89.57 | 89.83 |
| CK/541/18 | 96.12 | 94.67 | 95.49 | 94.88 | 97.56 | 96.80 | 96.43 | 96.35 |
| DK/01/22 | 88.24 | 87.76 | 87.58 | 87.31 | 90.00 | 90.00 | 89.46 | 89.43 |
Table 3 Nucleotide homology and amino acid homology of the hemagglutinin between five H9N2 subtype AIV isolates and vaccine strains %
| 毒株 Strain | 核苷酸同源性Nucleotide homology | 氨基酸同源性Amino acid homology | ||||||
|---|---|---|---|---|---|---|---|---|
| CK/SS/94 | CK/F/98 | CK/6/96 | CK/1/00 | CK/SS/94 | CK/F/98 | CK/6/96 | CK/1/00 | |
| CK/01/22 | 88.35 | 87.88 | 87.70 | 87.44 | 90.18 | 90.18 | 89.64 | 89.62 |
| QU/01/12 | 87.84 | 86.60 | 86.78 | 86.78 | 89.70 | 88.39 | 88.80 | 88.53 |
| QU/535/12 | 88.46 | 87.21 | 88.08 | 87.41 | 90.82 | 89.14 | 89.57 | 89.83 |
| CK/541/18 | 96.12 | 94.67 | 95.49 | 94.88 | 97.56 | 96.80 | 96.43 | 96.35 |
| DK/01/22 | 88.24 | 87.76 | 87.58 | 87.31 | 90.00 | 90.00 | 89.46 | 89.43 |
| 分离株 Strain | 裂解位点 Cleavage site | 左侧臂 Left side wall | 右侧臂 Right side wall | 受体结合位点 Receptor binding site | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 333-341 | 232-237 | 146-150 | 109 | 161 | 163 | 191 | 198 | 202 | 203 | 224 | |
| CK/GHA/17* | PARSSR↓GLF | NGLIGR | GTSKS | Y | W | T | H | A | L | Y | L |
| CK/BJ/94* | PARSSR↓GLF | NGQQGR | GTSKA | Y | W | T | N | V | L | Y | Q |
| CK/F/98* | PARSSR↓GLF | NGQQGR | GTSKA | Y | W | T | N | A | L | Y | Q |
| CK/SS/94* | PAGSSR↓GLF | NGQQGR | GTSKA | Y | W | T | N | A | L | Y | V |
| DK/Y280/97* | PARSSR↓GLF | NGLQGR | GTSKA | Y | W | T | N | T | L | Y | L |
| CK/43/02* | PARSSR↓GLF | NGLQGR | GTSRA | Y | W | T | N | A | L | Y | L |
| EN/01/2021* | PSRSSR↓GLF | NGLMGR | GTSNT | Y | W | N | N | V | L | Y | L |
| CK/D3/2022* | PSRSSR↓GLF | NGLMGR | GTSKT | Y | W | N | N | V | L | Y | L |
| CK/01/22 | PSRSSR↓GLF | NGLMGR | GTSTA | Y | W | T | N | T | L | Y | L |
| QU/01/12 | PARSSR↓GLF | NGQQGR | GTSIS | Y | W | T | H | A | L | Y | V |
| QU/535/12 | PARSSR↓GLF | NGQQGR | GTSIS | Y | W | T | H | T | L | Y | V |
| CK/541/18 | PARLSR↓GLF | NGQQGR | GTSKA | Y | W | T | N | T | L | Y | L |
| DK/01/22 | PSRSSR↓GLF | NGLMGR | GTSTA | Y | W | T | N | T | L | Y | L |
Table 4 Cleavage sites and receptor binding sites of hemagglutinin proteins of five isolates
| 分离株 Strain | 裂解位点 Cleavage site | 左侧臂 Left side wall | 右侧臂 Right side wall | 受体结合位点 Receptor binding site | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 333-341 | 232-237 | 146-150 | 109 | 161 | 163 | 191 | 198 | 202 | 203 | 224 | |
| CK/GHA/17* | PARSSR↓GLF | NGLIGR | GTSKS | Y | W | T | H | A | L | Y | L |
| CK/BJ/94* | PARSSR↓GLF | NGQQGR | GTSKA | Y | W | T | N | V | L | Y | Q |
| CK/F/98* | PARSSR↓GLF | NGQQGR | GTSKA | Y | W | T | N | A | L | Y | Q |
| CK/SS/94* | PAGSSR↓GLF | NGQQGR | GTSKA | Y | W | T | N | A | L | Y | V |
| DK/Y280/97* | PARSSR↓GLF | NGLQGR | GTSKA | Y | W | T | N | T | L | Y | L |
| CK/43/02* | PARSSR↓GLF | NGLQGR | GTSRA | Y | W | T | N | A | L | Y | L |
| EN/01/2021* | PSRSSR↓GLF | NGLMGR | GTSNT | Y | W | N | N | V | L | Y | L |
| CK/D3/2022* | PSRSSR↓GLF | NGLMGR | GTSKT | Y | W | N | N | V | L | Y | L |
| CK/01/22 | PSRSSR↓GLF | NGLMGR | GTSTA | Y | W | T | N | T | L | Y | L |
| QU/01/12 | PARSSR↓GLF | NGQQGR | GTSIS | Y | W | T | H | A | L | Y | V |
| QU/535/12 | PARSSR↓GLF | NGQQGR | GTSIS | Y | W | T | H | T | L | Y | V |
| CK/541/18 | PARLSR↓GLF | NGQQGR | GTSKA | Y | W | T | N | T | L | Y | L |
| DK/01/22 | PSRSSR↓GLF | NGLMGR | GTSTA | Y | W | T | N | T | L | Y | L |
| 分离株 Strain | 糖基化位点Glycosylation site | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 29 | 82 | 141 | 206 | 218 | 298 | 305 | 313 | 492 | 551 | |
| CK/GHA/17* | NSTE | NPSC | NVTY | NSTL | NISK | NGTY | NGSC | |||
| CK/BJ/94* | NSTE | NPSC | NVTY | NRTF | NTTL | NVSK | NGTY | NGSC | ||
| CK/F/98* | NSTE | NPSC | NVSY | NTTL | NVSK | NGTY | NGSC | |||
| CK/SS/94* | NSTE | NPSC | NVSY | NRTF | NTTL | NVSK | NGTY | NGSC | ||
| Dk/Y280/97* | NSTE | NPSC | NVSY | NRTF | NTTL | NVSK | NGTY | |||
| CK/43/02* | NSTE | NPSC | NVSY | NRTF | NTTL | NVSK | NGTY | NGSC | ||
| EN/01/2021* | NSTE | NPSC | NVSY | NTTL | NVSK | NCSK | NGTY | NGSC | ||
| CK/D3/2022* | NSTE | NPSC | NVSY | NTTL | NVSK | NCSK | NGTY | NGSC | ||
| CK/01/22 | NSTE | NPSC | NVSY | NTTL | NVSK | NCSK | NGTY | NGSC | ||
| QU/01/12 | NSTE | NPSC | NVSY | NDTT | NRTF | NSTL | NISK | NGTY | ||
| QU/535/12 | NSTE | NPSC | NVSY | NDTT | NRTF | NSTL | NISK | NGTY | ||
| CK/541/18 | NSTE | NPSC | NVSY | NRTF | NTTL | NVSK | NGTY | |||
| DK/01/22 | NSTE | NPSC | NVSY | NTTL | NVSK | NCSK | NGTY | NGSC | ||
Table 5 Potential glycosylation sites in hemagglutinin of five H9N2 subtype AIV isolates
| 分离株 Strain | 糖基化位点Glycosylation site | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 29 | 82 | 141 | 206 | 218 | 298 | 305 | 313 | 492 | 551 | |
| CK/GHA/17* | NSTE | NPSC | NVTY | NSTL | NISK | NGTY | NGSC | |||
| CK/BJ/94* | NSTE | NPSC | NVTY | NRTF | NTTL | NVSK | NGTY | NGSC | ||
| CK/F/98* | NSTE | NPSC | NVSY | NTTL | NVSK | NGTY | NGSC | |||
| CK/SS/94* | NSTE | NPSC | NVSY | NRTF | NTTL | NVSK | NGTY | NGSC | ||
| Dk/Y280/97* | NSTE | NPSC | NVSY | NRTF | NTTL | NVSK | NGTY | |||
| CK/43/02* | NSTE | NPSC | NVSY | NRTF | NTTL | NVSK | NGTY | NGSC | ||
| EN/01/2021* | NSTE | NPSC | NVSY | NTTL | NVSK | NCSK | NGTY | NGSC | ||
| CK/D3/2022* | NSTE | NPSC | NVSY | NTTL | NVSK | NCSK | NGTY | NGSC | ||
| CK/01/22 | NSTE | NPSC | NVSY | NTTL | NVSK | NCSK | NGTY | NGSC | ||
| QU/01/12 | NSTE | NPSC | NVSY | NDTT | NRTF | NSTL | NISK | NGTY | ||
| QU/535/12 | NSTE | NPSC | NVSY | NDTT | NRTF | NSTL | NISK | NGTY | ||
| CK/541/18 | NSTE | NPSC | NVSY | NRTF | NTTL | NVSK | NGTY | |||
| DK/01/22 | NSTE | NPSC | NVSY | NTTL | NVSK | NCSK | NGTY | NGSC | ||
| 分离株 Strain | 抗原性位点Antigenic site | ||||||
|---|---|---|---|---|---|---|---|
| 181 | 234 | 280 | 281 | 285 | 296 | 334 | |
| CK/GHA/17* | G | L | D | L | N | G | A |
| CK/BJ/94* | G | Q | D | L | N | G | A |
| CK/F/98* | G | Q | D | L | N | G | A |
| CK/SS/94* | G | Q | D | L | N | G | A |
| Dk/Y280/97* | G | L | D | L | N | G | A |
| CK/43/02* | G | L | D | L | N | G | A |
| EN/01/2021* | G | L | D | L | S | G | S |
| CK/D3/2022* | G | L | D | L | S | G | S |
| CK/01/22 | G | L | D | L | S | G | S |
| QU/01/12 | G | Q | D | L | N | G | A |
| QU/535/12 | G | Q | D | L | N | G | A |
| CK/541/18 | G | Q | D | L | N | G | A |
| DK/01/22 | G | L | D | L | S | G | S |
Table 6 The hemagglutinin antigenic loci of five H9N2 subtype AIV isolates
| 分离株 Strain | 抗原性位点Antigenic site | ||||||
|---|---|---|---|---|---|---|---|
| 181 | 234 | 280 | 281 | 285 | 296 | 334 | |
| CK/GHA/17* | G | L | D | L | N | G | A |
| CK/BJ/94* | G | Q | D | L | N | G | A |
| CK/F/98* | G | Q | D | L | N | G | A |
| CK/SS/94* | G | Q | D | L | N | G | A |
| Dk/Y280/97* | G | L | D | L | N | G | A |
| CK/43/02* | G | L | D | L | N | G | A |
| EN/01/2021* | G | L | D | L | S | G | S |
| CK/D3/2022* | G | L | D | L | S | G | S |
| CK/01/22 | G | L | D | L | S | G | S |
| QU/01/12 | G | Q | D | L | N | G | A |
| QU/535/12 | G | Q | D | L | N | G | A |
| CK/541/18 | G | Q | D | L | N | G | A |
| DK/01/22 | G | L | D | L | S | G | S |
| [1] | HOMME P J, EASTERDAY B C. Avian influenza virus infections. I. characteristics of influenza A-Turkey-Wisconsin-1966 virus[J]. Avian Diseases, 1970, 14(1): 66. |
| [2] | DABRERA G. H5 and H9 avian influenza-potential re-emergent zoonotic threats to humans[J]. Current Opinion in Infectious Diseases, 2024, 37(5): 431-435. |
| [3] | GU M, XU L J, WANG X Q, et al. Current situation of H9N2 subtype avian influenza in China[J]. Veterinary Research, 2017, 48(1): 49. |
| [4] | 刘照生, 伏晓庆, 罗春蕊, 等. 云南省2023年活禽市场8株H9N2亚型禽流感病毒全基因组序列分析[J]. 中国热带医学, 2025, 25(3): 350-357. |
| LIU Z S, FU X Q, LUO C R, et al. Whole genomic analysis of 8 strains of H9N2 subtype avian influenza virus isolates from live poultry markets in Yunnan, 2023[J]. China Tropical Medicine, 2025, 25(3): 350-357. (in Chinese with English abstract) | |
| [5] | PENG Q Q, ZHU R, WANG X B, et al. Impact of the variations in potential glycosylation sites of the hemagglutinin of H9N2 influenza virus[J]. Virus Genes, 2019, 55(2): 182-190. |
| [6] | HUANG X Y, YIN G H, CAI Y Q, et al. Identification of unique and conserved neutralizing epitopes of vestigial esterase domain in HA protein of the H9N2 subtype of avian influenza virus[J]. Viruses, 2022, 14(12): 2739. |
| [7] | 曾庆航, 刘杨, 孙敏华, 等. 广东地区H9N2亚型禽流感病毒HA和NA基因生物学特征分析[J]. 广东农业科学, 2023, 50(5): 103-111. |
| ZENG Q H, LIU Y, SUN M H, et al. Biological characteristic of HA and NA genes of H9N2 avian influenza virus in Guangdong Province[J]. Guangdong Agricultural Sciences, 2023, 50(5): 103-111. (in Chinese with English abstract) | |
| [8] | LIU Q Z, ZHAO L C, GUO Y N, et al. Antigenic evolution characteristics and immunological evaluation of H9N2 avian influenza viruses from 1994-2019 in China[J]. Viruses, 2022, 14(4): 726. |
| [9] | KIDO H, YOKOGOSHI Y, SAKAI K, et al. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells: a possible activator of the viral fusion glycoprotein[J]. Journal of Biological Chemistry, 1992, 267(19): 13573-13579. |
| [10] | GAO Y W, ZHANG Y, SHINYA K, et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host[J]. PLoS Pathogens, 2009, 5(12): e1000709. |
| [11] | MATROSOVICH M N, KRAUSS S, WEBSTER R G. H9N2 influenza a viruses from poultry in Asia have human virus-like receptor specificity[J]. Virology, 2001, 281(2): 156-162. |
| [12] | 杨婧, 刘宇卓, 赵冬敏, 等. 2013年苏皖地区4株H9N2亚型禽流感病毒HA、NA基因的遗传演化分析[J]. 浙江农业学报, 2015, 27(11): 1896-1902. |
| YANG J, LIU Y Z, ZHAO D M, et al. Phylogenetic analysis of the HA and NA genes of four strains of H9N2 subtype of avian influenza virus isolated from Jiangsu and Anhui Province in 2013[J]. Acta Agriculturae Zhejiangensis, 2015, 27(11): 1896-1902. (in Chinese with English abstract) | |
| [13] | AYTAY S, SCHULZE I T. Single amino acid substitutions in the hemagglutinin can alter the host range and receptor binding properties of H1 strains of influenza A virus[J]. Journal of Virology, 1991, 65(6): 3022-3028. |
| [14] | 邢燕茹, 祝宇翔, 范春燕, 等. 6株H9N2亚型禽流感病毒分子特征及遗传演化分析[J]. 畜牧与兽医, 2024, 56(2): 89-97. |
| XING Y R, ZHU Y X, FAN C Y, et al. Molecular characterization and genetic evolution of six H9N2 subtype avian influenza virus strains[J]. Animal Husbandry & Veterinary Medicine, 2024, 56(2): 89-97. (in Chinese with English abstract) |
| [1] | QI Yingji, LIU Shuya, LAN Hanyun, WANG Xuemei, SUN Tiantian, LI Shuhan, LI Qiunan, HUANG Xiaoli, GENG Yi, CHEN Defang, OUYANG Ping. Isolation, identification and genome-wide analysis of Spring viremia of carp virus in Sichuan Province of China [J]. Acta Agriculturae Zhejiangensis, 2025, 37(7): 1430-1440. |
| [2] | NI Zheng, LU Lei, ZHU Yinchu, CHEN Liu, LI Xinji, YE Weicheng, YUN Tao, HUA Jionggang, FU Yuan, ZHANG Cun. Epidemiological investigation and evaluation of purification strategies for avian leukosis in local chicken breeds in Zhejiang Province of China [J]. Acta Agriculturae Zhejiangensis, 2025, 37(4): 790-799. |
| [3] | YANG Xiaoyu, MA Zhihui, WEI Qing, NIU Zhipeng, CHEN Anqi, HU Zhengchong, WANG Linsheng. Preliminary mapping of a wheat awn length gene and prediction of candidate genes [J]. Acta Agriculturae Zhejiangensis, 2025, 37(1): 14-23. |
| [4] | CAI Shiyi, YU Huifang, WANG Jiansheng, ZHU Biao, SHEN Yusen, GU Honghui, SHENG Xiaoguang. Major gene plus polygene inheritance analysis of curd sitting height in cauliflower [J]. Acta Agriculturae Zhejiangensis, 2024, 36(3): 527-533. |
| [5] | SHOU Weisong, HE Yanjun, SHEN Jia, XU Xinyang. Genome-wide identification and bioinformatics analysis of SWEET gene family in melon [J]. Acta Agriculturae Zhejiangensis, 2023, 35(7): 1591-1603. |
| [6] | JIA Beiping, LYU Xuan, YANG Qing, WANG Yinan, LI Wanxiao, XIE Xindi, ZHU Yingqi, WANG Bei, YIN Dongdong, ZHANG Yunkai, WANG Qing, WANG Guijun. Isolation, identification and genetic evolution analysis of novel goose astrovirus in Anhui Province, China [J]. Acta Agriculturae Zhejiangensis, 2023, 35(5): 1048-1057. |
| [7] | LANG Chunxiu, LIU Renhu, ZHENG Tao, WANG Fulin, SHI Jianghua, HU Zhanghua, WU Guanting. New dwarf mutants of oilseed rape (Brassica napus L.) induced by chemical mutagenesis [J]. Acta Agriculturae Zhejiangensis, 2023, 35(11): 2516-2524. |
| [8] | WEI Haizhong, PAN Liqin, TANG Ziyi, TIAN Shengye, HE Haiye, YIN Longfei, ZHENG Dewei, ZHANG Huijuan, JIANG Ming. Isolation and expression analysis of a SKIP gene from Brassica oleracea var. italica [J]. Acta Agriculturae Zhejiangensis, 2022, 34(5): 966-973. |
| [9] | LIU Tongjin, XU Mingjie, WANG Jinglei, LIU Liangfeng, CUI Qunxiang, BAO Chonglai, WANG Changyi. Genome-wide identification and expression analysis of ALMT gene family in radish [J]. Acta Agriculturae Zhejiangensis, 2022, 34(4): 746-755. |
| [10] | ZHAO Hongxi, LIU Jibing. Infection investigation and genetic evolution analysis of dairy cow coccidiosis in parts of Ningxia [J]. Acta Agriculturae Zhejiangensis, 2021, 33(8): 1379-1384. |
| [11] | ZHAO Guofu, YAN Yaqin, WANG Jinglei, WEI Qingzhen, BAO Chonglai. Genome-wide identification and expression analysis of LOX gene family in eggplant (Solanum melongena) [J]. Acta Agriculturae Zhejiangensis, 2021, 33(6): 1025-1034. |
| [12] | ZHAO Ke, LI Qiurong, HOU Lu, BAI Yaobo, JIANG Liling, WEI Youhai, GUO Qingyun. Genetic analysis of stripe rust resistance genes in 2 spring wheat germplasm resources at adult stage [J]. Acta Agriculturae Zhejiangensis, 2021, 33(4): 595-601. |
| [13] | JI Kaiyuan, QIU Yueyang, CHENG Ao, JIANG Shudong, PENG Mengling. Genetic diversity of Canine parvovirus in Hefei, Anhui Province from 2018 to 2019 [J]. Acta Agriculturae Zhejiangensis, 2021, 33(10): 1817-1825. |
| [14] | PU Lusha, SU Shibo, CHEN Xiaohan, ZHAO Lili, CHEN Hongyan. Prokaryotic expression and phylogenetic analysis of ORF2 gene of goose astrovirus [J]. , 2020, 32(5): 789-797. |
| [15] | JIA Xiaoping, WANG Zhenshan, ZHU Xuehai, YANG Dezhi, KOU Shujun, LIU Xingxing. Genetic analysis of dwarf gene for dwarf mutant“819” in Panicum miliaceum L. [J]. , 2020, 32(1): 20-27. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||