浙江农业学报 ›› 2025, Vol. 37 ›› Issue (10): 2087-2103.DOI: 10.3969/j.issn.1004-1524.20241128
宽叶苔草WRKY家族成员生物信息学分析与耐旱基因挖掘
崔博文( ), 张思懿, 王佳玲, 王竞红, 蔺吉祥, 杨青杰(
), 张思懿, 王佳玲, 王竞红, 蔺吉祥, 杨青杰( )
)
- 东北林业大学 园林学院,黑龙江 哈尔滨 150040
-
收稿日期:2024-12-30出版日期:2025-10-25发布日期:2025-11-13 -
作者简介:崔博文(1999—),男,黑龙江哈尔滨人,硕士,研究方向为园林植物栽培育种。E-mail:cuibowen1999@163.com -
通讯作者:杨青杰,E-mail:qingjieyang@nefu.edu.cn -
基金资助:国家自然科学基金(32072666);国家重点研发计划青年科学家项目(2023YFF1305800)
Bioinformatics analysis and drought-tolerant gene mining of WRKY family members in Carex siderosticta
CUI Bowen( ), ZHANG Siyi, WANG Jialing, WANG Jinghong, LIN Jixiang, YANG Qingjie(
), ZHANG Siyi, WANG Jialing, WANG Jinghong, LIN Jixiang, YANG Qingjie( )
)
- College of Landscape Architecture, Northeast Forestry University, Harbin 150040, China
-
Received:2024-12-30Online:2025-10-25Published:2025-11-13
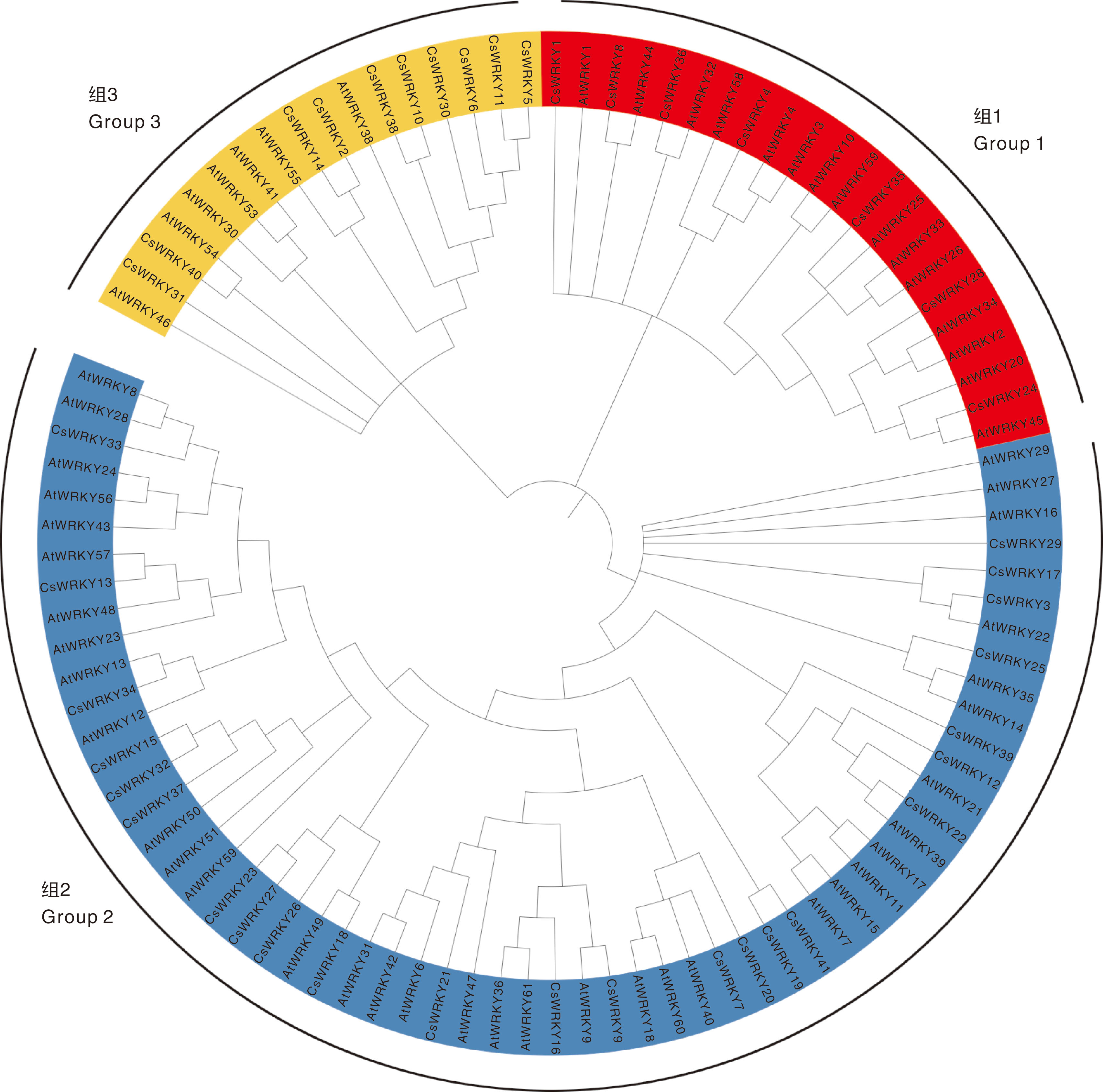
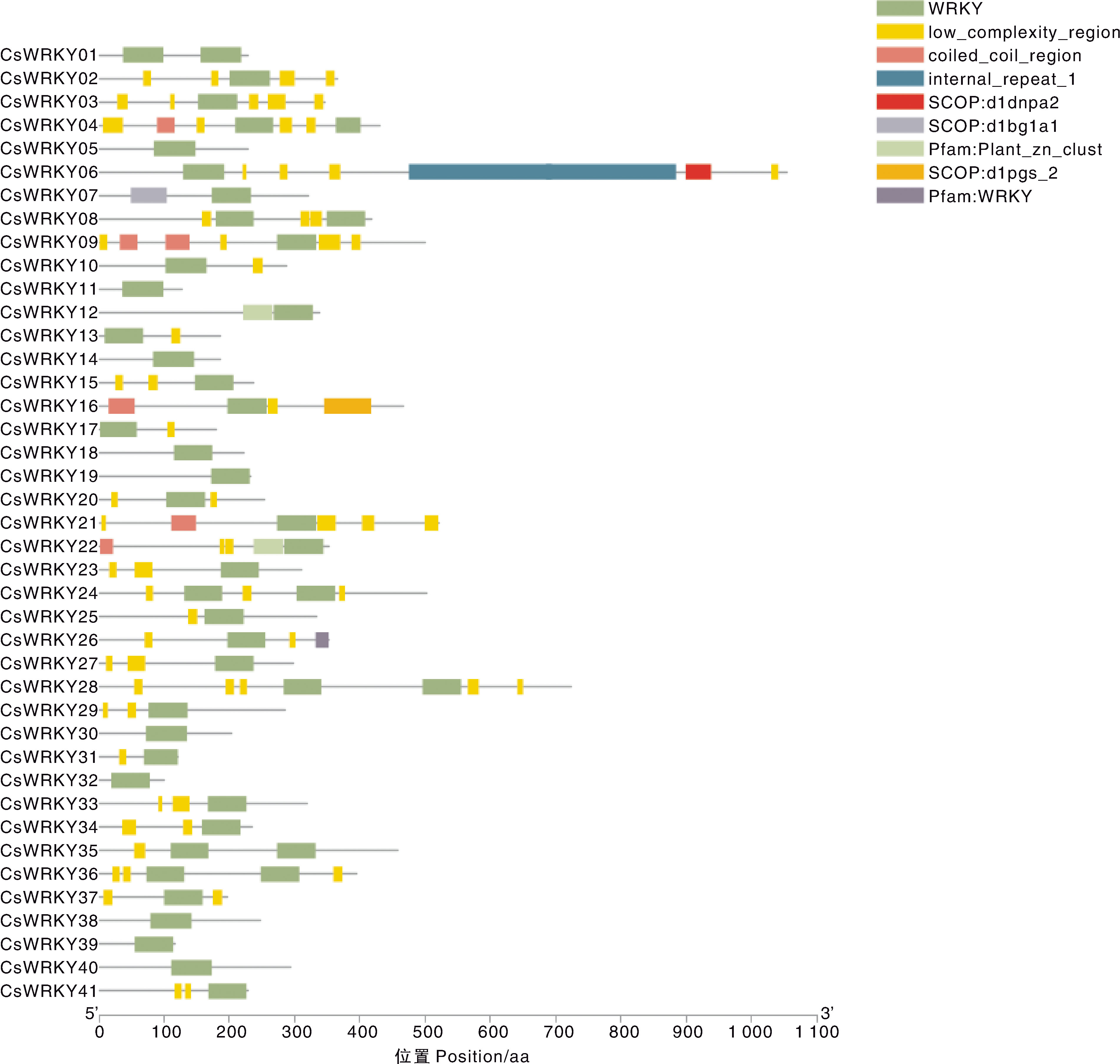
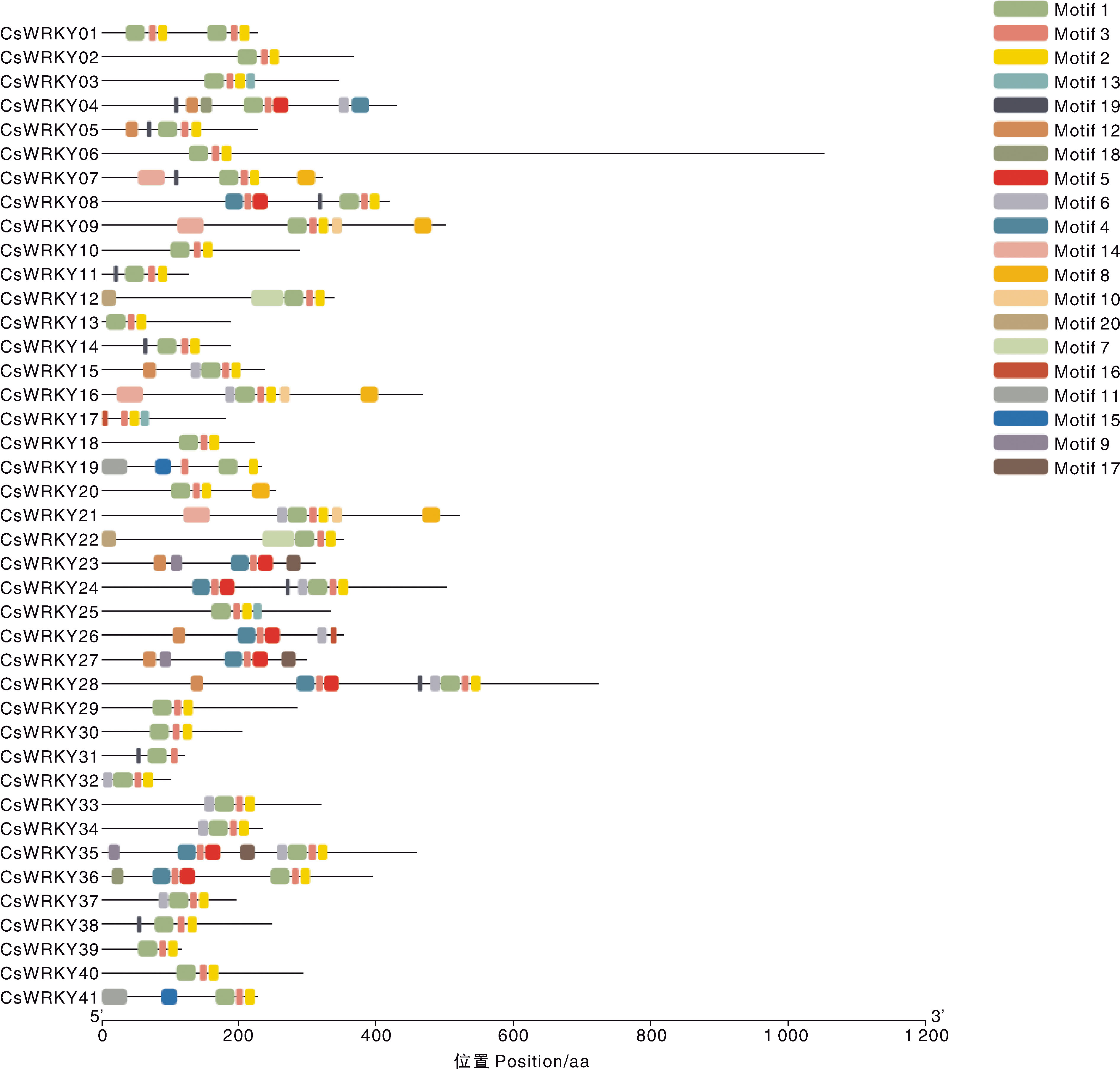
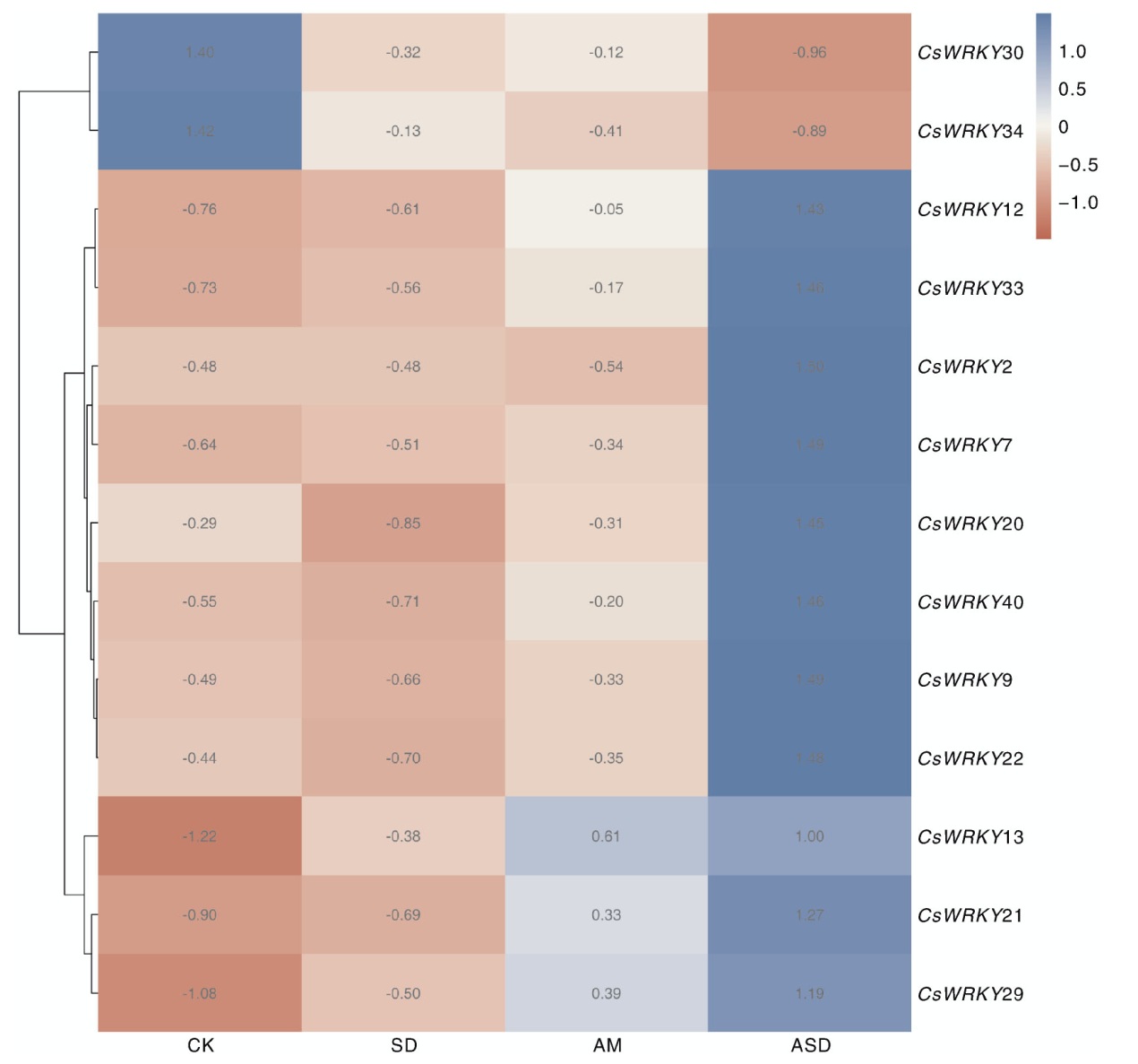
摘要: 为深入研究WRKY转录因子在干旱和丛枝菌根(AM)处理下的生物学功能,本研究对接种AM真菌和干旱胁迫的宽叶苔草(Carex siderosticta)进行转录组学测序,鉴定宽叶苔草WRKY(CsWRKY)基因家族成员,分析其理化性质、蛋白质二级结构、保守基序与表达模式等,结合qRT-PCR验证,挖掘与干旱胁迫相关的候选WRKY基因。结果表明,共获得41个CsWRKY基因家族成员,划分为3类,组1有7个,组2有24个,组3有10个。CsWRKY基因家族成员的氨基酸数为99~1 052,理论等电点为4.83~9.84,亲水性系数均为负值,表明均为亲水蛋白。GO注释富集于转录调控、代谢过程调控、生物合成过程调控3大类功能。KEGG通路主要富集于植物MAPK信号通路、植物-病原互作、转录因子3个通路。接种AM后,26个CsWRKY基因表达上调,12个下调。结合转录组数据与同源比对,筛选出13个可能参与干旱响应的CsWRKY候选基因。qRT-PCR分析表明,10个候选基因受干旱胁迫诱导,可能发挥正调控作用;另3个候选基因受干旱胁迫抑制,可能发挥负调控作用。本研究为解析宽叶苔草抗旱机制提供了候选基因资源与理论依据。
中图分类号:
引用本文
崔博文, 张思懿, 王佳玲, 王竞红, 蔺吉祥, 杨青杰. 宽叶苔草WRKY家族成员生物信息学分析与耐旱基因挖掘[J]. 浙江农业学报, 2025, 37(10): 2087-2103.
CUI Bowen, ZHANG Siyi, WANG Jialing, WANG Jinghong, LIN Jixiang, YANG Qingjie. Bioinformatics analysis and drought-tolerant gene mining of WRKY family members in Carex siderosticta[J]. Acta Agriculturae Zhejiangensis, 2025, 37(10): 2087-2103.
| 基因名称 Gene name | 正向引物序列 Forward primer sequence(5'-3') | 反向引物序列 Reverse primer sequence(5'-3') |
|---|---|---|
| CsEF-1α | AGTGGTTATCGGTCACGTCG | GTTCATCTCAGCGGCTTCCT |
| CsWRKY2 | AATGTGGAGGTTGGTGGGAC | GGACCCGTTGCATCACTCTT |
| CsWRKY7 | GGTTCAAAGAAGCGCGGAAG | AAGTCGGTTTTCGTGCAAGC |
| CsWRKY9 | GCAATCCGAGATGGATCGGT | GCTCTTTTTGCTTCCCGGTG |
| CsWRKY12 | AATGCAGCGGAGTGAAAGGT | TGGTTGTGCTCGCCTTCATA |
| CsWRKY13 | CCAGCTCAGGTTGGTGTTTTG | CCCTCATCCATCCGGCTTAC |
| CsWRKY20 | ACAATGGCTCCAGCAAGACA | ATGATCGGCCTGTGGTTCAG |
| CsWRKY21 | CCACCATGAGGAAAGCTCGT | CTTCAGCACACCTTTGCACC |
| CsWRKY22 | ATGCCTCGTCGTGTTCATCT | CTTGACTTGCTGGAGGTGGT |
| CsWRKY29 | CTAATCACTGCGCCTGACGA | CGTCTTCCCCCGCTTGTAAT |
| CsWRKY30 | GCAGCCACAAAACAAGTCCA | CTGACATTGGTGTCGAGTTGC |
| CsWRKY33 | TTCTTCGTCGTGTGAGGCAA | GTGGCTCACGTTGCCTTTTC |
| CsWRKY34 | ACGGTCAGAAGGTCGTCAAG | GCATGTCGTCCCTCGTATGT |
| CsWRKY40 | GAGGAAGGGTTTTGACGGGT | CCTGGTCGAATTTGTGCGTG |
表1 CsWRKY基因的qRT-PCR引物序列
Table 1 qRT-PCR primer sequences for CsWRKY gene
| 基因名称 Gene name | 正向引物序列 Forward primer sequence(5'-3') | 反向引物序列 Reverse primer sequence(5'-3') |
|---|---|---|
| CsEF-1α | AGTGGTTATCGGTCACGTCG | GTTCATCTCAGCGGCTTCCT |
| CsWRKY2 | AATGTGGAGGTTGGTGGGAC | GGACCCGTTGCATCACTCTT |
| CsWRKY7 | GGTTCAAAGAAGCGCGGAAG | AAGTCGGTTTTCGTGCAAGC |
| CsWRKY9 | GCAATCCGAGATGGATCGGT | GCTCTTTTTGCTTCCCGGTG |
| CsWRKY12 | AATGCAGCGGAGTGAAAGGT | TGGTTGTGCTCGCCTTCATA |
| CsWRKY13 | CCAGCTCAGGTTGGTGTTTTG | CCCTCATCCATCCGGCTTAC |
| CsWRKY20 | ACAATGGCTCCAGCAAGACA | ATGATCGGCCTGTGGTTCAG |
| CsWRKY21 | CCACCATGAGGAAAGCTCGT | CTTCAGCACACCTTTGCACC |
| CsWRKY22 | ATGCCTCGTCGTGTTCATCT | CTTGACTTGCTGGAGGTGGT |
| CsWRKY29 | CTAATCACTGCGCCTGACGA | CGTCTTCCCCCGCTTGTAAT |
| CsWRKY30 | GCAGCCACAAAACAAGTCCA | CTGACATTGGTGTCGAGTTGC |
| CsWRKY33 | TTCTTCGTCGTGTGAGGCAA | GTGGCTCACGTTGCCTTTTC |
| CsWRKY34 | ACGGTCAGAAGGTCGTCAAG | GCATGTCGTCCCTCGTATGT |
| CsWRKY40 | GAGGAAGGGTTTTGACGGGT | CCTGGTCGAATTTGTGCGTG |
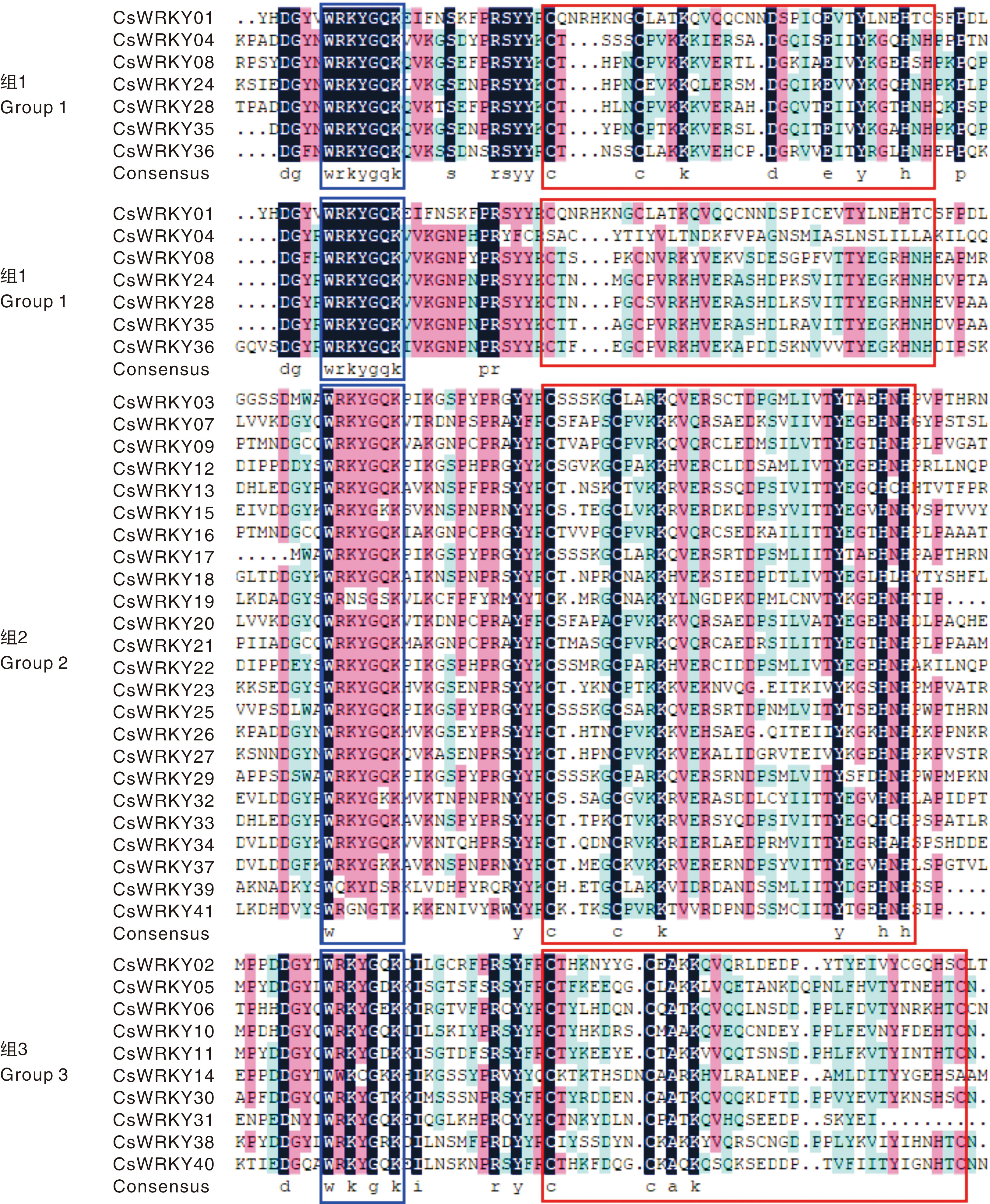
图1 CsWRKY家族保守结构域序列比对 蓝色框内为WRKY结构域,红色框内为锌指结构。
Fig.1 Sequences alignment of conserved domains in the CsWRKY family The WRKY domain is located within the blue box, the zinc finger structure is enclosed within the red box.
| 蛋白质名称 Protein name | 蛋白质ID Protein ID | 氨基酸数量 Number of amino acids | 分子量 Molecular weight/u | 理论等电点 Theoretical pI | 脂肪系数 Aliphatic index | 亲水性指数 Hydropathicity index | 可靠性指数 Reliable index |
|---|---|---|---|---|---|---|---|
| CsWRKY1 | TRINITY_DN25341_c0_g1.p1 | 227 | 25 859.92 | 9.13 | 53.66 | -0.861 | 2.713 |
| CsWRKY2 | TRINITY_DN71036_c0_g1.p1 | 365 | 39 547.20 | 6.26 | 71.32 | -0.387 | 3.220 |
| CsWRKY3 | TRINITY_DN24014_c0_g1.p1 | 345 | 37 300.06 | 5.16 | 72.09 | -0.506 | 4.106 |
| CsWRKY4 | TRINITY_DN21305_c0_g1.p1 | 429 | 46 690.97 | 8.43 | 59.79 | -0.805 | 4.541 |
| CsWRKY5 | TRINITY_DN23710_c0_g1.p1 | 227 | 25 812.35 | 5.98 | 64.89 | -0.558 | 2.769 |
| CsWRKY6 | TRINITY_DN8042_c0_g1.p1 | 1 052 | 114 418.31 | 6.16 | 60.91 | -0.774 | 3.934 |
| CsWRKY7 | TRINITY_DN685_c1_g3.p1 | 320 | 36 371.66 | 7.01 | 63.09 | -0.854 | 4.610 |
| CsWRKY8 | TRINITY_DN13228_c0_g1.p1 | 418 | 46 029.94 | 8.90 | 48.71 | -0.876 | 4.399 |
| CsWRKY9 | TRINITY_DN50400_c0_g1.p1 | 499 | 55 216.80 | 5.22 | 58.10 | -0.941 | 4.819 |
| CsWRKY10 | TRINITY_DN2138_c0_g1.p1 | 287 | 32 783.87 | 6.02 | 72.02 | -0.653 | 4.224 |
| CsWRKY11 | TRINITY_DN35748_c0_g2.p1 | 127 | 15 061.14 | 9.35 | 60.55 | -0.846 | 2.472 |
| CsWRKY12 | TRINITY_DN1223_c0_g2.p1 | 337 | 37 461.27 | 9.54 | 64.51 | -0.785 | 4.270 |
| CsWRKY13 | TRINITY_DN920_c0_g1.p1 | 186 | 20 375.75 | 8.99 | 63.44 | -0.637 | 3.681 |
| CsWRKY14 | TRINITY_DN26245_c0_g2.p1 | 186 | 20 497.80 | 8.25 | 48.82 | -0.712 | 4.341 |
| CsWRKY15 | TRINITY_DN10828_c1_g1.p1 | 236 | 25 423.32 | 5.94 | 54.19 | -0.586 | 3.227 |
| CsWRKY16 | TRINITY_DN52127_c0_g1.p1 | 466 | 51 580.45 | 8.02 | 60.11 | -0.789 | 4.775 |
| CsWRKY17 | TRINITY_DN10234_c0_g1.p1 | 180 | 19 948.94 | 4.83 | 49.39 | -0.923 | 3.832 |
| CsWRKY18 | TRINITY_DN88517_c0_g1.p1 | 222 | 25 105.91 | 6.66 | 61.53 | -0.936 | 4.163 |
| CsWRKY19 | TRINITY_DN60019_c0_g1.p1 | 232 | 26 356.25 | 6.00 | 73.92 | -0.487 | 4.009 |
| CsWRKY20 | TRINITY_DN5707_c0_g1.p1 | 253 | 27 820.48 | 8.97 | 66.68 | -0.733 | 4.411 |
| CsWRKY21 | TRINITY_DN238_c2_g1.p1 | 520 | 55 805.56 | 5.75 | 60.35 | -0.569 | 4.501 |
| CsWRKY22 | TRINITY_DN12424_c0_g1.p1 | 352 | 39 065.32 | 9.84 | 66.22 | -0.782 | 4.599 |
| CsWRKY23 | TRINITY_DN4346_c0_g1.p1 | 309 | 33 839.20 | 7.73 | 52.72 | -0.891 | 4.207 |
| CsWRKY24 | TRINITY_DN8931_c0_g1.p1 | 502 | 54 633.32 | 6.44 | 57.45 | -0.878 | 3.872 |
| CsWRKY25 | TRINITY_DN32124_c3_g1.p1 | 333 | 35 836.41 | 7.66 | 64.23 | -0.535 | 3.024 |
| CsWRKY26 | TRINITY_DN26434_c0_g1.p1 | 352 | 38 775.68 | 6.14 | 50.99 | -0.985 | 4.718 |
| CsWRKY27 | TRINITY_DN714_c2_g1.p1 | 298 | 32 980.16 | 6.20 | 48.12 | -0.912 | 3.346 |
| CsWRKY28 | TRINITY_DN3255_c1_g1.p1 | 722 | 78 422.37 | 5.95 | 48.49 | -0.854 | 3.944 |
| CsWRKY29 | TRINITY_DN4758_c0_g1.p1 | 284 | 30 970.33 | 5.31 | 60.11 | -0.711 | 3.870 |
| CsWRKY30 | TRINITY_DN82013_c0_g1.p1 | 204 | 23 014.52 | 7.04 | 47.79 | -0.749 | 3.481 |
| CsWRKY31 | TRINITY_DN54950_c0_g1.p1 | 121 | 13 931.73 | 9.25 | 62.07 | -1.107 | 2.837 |
| CsWRKY32 | TRINITY_DN44830_c0_g1.p1 | 99 | 11 695.20 | 9.23 | 58.08 | -0.890 | 2.449 |
| CsWRKY33 | TRINITY_DN4854_c0_g1.p1 | 318 | 35 704.86 | 7.11 | 55.50 | -0.876 | 4.841 |
| CsWRKY34 | TRINITY_DN28623_c0_g1.p1 | 234 | 26 682.65 | 9.28 | 51.62 | -0.981 | 4.762 |
| CsWRKY35 | TRINITY_DN15_c2_g1.p1 | 458 | 51 061.95 | 5.85 | 47.53 | -0.967 | 4.421 |
| CsWRKY36 | TRINITY_DN7274_c0_g1.p1 | 394 | 43 242.46 | 8.51 | 57.13 | -0.808 | 4.270 |
| CsWRKY37 | TRINITY_DN9415_c1_g1.p1 | 195 | 22 127.53 | 7.64 | 53.49 | -0.791 | 3.354 |
| CsWRKY38 | TRINITY_DN87366_c0_g1.p1 | 247 | 28 132.69 | 5.20 | 75.75 | -0.517 | 2.689 |
| CsWRKY39 | TRINITY_DN50547_c0_g2.p1 | 115 | 13 177.86 | 9.23 | 58.52 | -0.937 | 2.686 |
| CsWRKY40 | TRINITY_DN5168_c0_g1.p1 | 293 | 32 279.70 | 5.58 | 70.31 | -0.705 | 3.545 |
| CsWRKY41 | TRINITY_DN3189_c2_g1.p1 | 227 | 25 865.76 | 8.86 | 78.06 | -0.437 | 1.947 |
表2 CsWRKY转录因子的理化性质
Table 2 Physicochemical properties of CsWRKY proteins
| 蛋白质名称 Protein name | 蛋白质ID Protein ID | 氨基酸数量 Number of amino acids | 分子量 Molecular weight/u | 理论等电点 Theoretical pI | 脂肪系数 Aliphatic index | 亲水性指数 Hydropathicity index | 可靠性指数 Reliable index |
|---|---|---|---|---|---|---|---|
| CsWRKY1 | TRINITY_DN25341_c0_g1.p1 | 227 | 25 859.92 | 9.13 | 53.66 | -0.861 | 2.713 |
| CsWRKY2 | TRINITY_DN71036_c0_g1.p1 | 365 | 39 547.20 | 6.26 | 71.32 | -0.387 | 3.220 |
| CsWRKY3 | TRINITY_DN24014_c0_g1.p1 | 345 | 37 300.06 | 5.16 | 72.09 | -0.506 | 4.106 |
| CsWRKY4 | TRINITY_DN21305_c0_g1.p1 | 429 | 46 690.97 | 8.43 | 59.79 | -0.805 | 4.541 |
| CsWRKY5 | TRINITY_DN23710_c0_g1.p1 | 227 | 25 812.35 | 5.98 | 64.89 | -0.558 | 2.769 |
| CsWRKY6 | TRINITY_DN8042_c0_g1.p1 | 1 052 | 114 418.31 | 6.16 | 60.91 | -0.774 | 3.934 |
| CsWRKY7 | TRINITY_DN685_c1_g3.p1 | 320 | 36 371.66 | 7.01 | 63.09 | -0.854 | 4.610 |
| CsWRKY8 | TRINITY_DN13228_c0_g1.p1 | 418 | 46 029.94 | 8.90 | 48.71 | -0.876 | 4.399 |
| CsWRKY9 | TRINITY_DN50400_c0_g1.p1 | 499 | 55 216.80 | 5.22 | 58.10 | -0.941 | 4.819 |
| CsWRKY10 | TRINITY_DN2138_c0_g1.p1 | 287 | 32 783.87 | 6.02 | 72.02 | -0.653 | 4.224 |
| CsWRKY11 | TRINITY_DN35748_c0_g2.p1 | 127 | 15 061.14 | 9.35 | 60.55 | -0.846 | 2.472 |
| CsWRKY12 | TRINITY_DN1223_c0_g2.p1 | 337 | 37 461.27 | 9.54 | 64.51 | -0.785 | 4.270 |
| CsWRKY13 | TRINITY_DN920_c0_g1.p1 | 186 | 20 375.75 | 8.99 | 63.44 | -0.637 | 3.681 |
| CsWRKY14 | TRINITY_DN26245_c0_g2.p1 | 186 | 20 497.80 | 8.25 | 48.82 | -0.712 | 4.341 |
| CsWRKY15 | TRINITY_DN10828_c1_g1.p1 | 236 | 25 423.32 | 5.94 | 54.19 | -0.586 | 3.227 |
| CsWRKY16 | TRINITY_DN52127_c0_g1.p1 | 466 | 51 580.45 | 8.02 | 60.11 | -0.789 | 4.775 |
| CsWRKY17 | TRINITY_DN10234_c0_g1.p1 | 180 | 19 948.94 | 4.83 | 49.39 | -0.923 | 3.832 |
| CsWRKY18 | TRINITY_DN88517_c0_g1.p1 | 222 | 25 105.91 | 6.66 | 61.53 | -0.936 | 4.163 |
| CsWRKY19 | TRINITY_DN60019_c0_g1.p1 | 232 | 26 356.25 | 6.00 | 73.92 | -0.487 | 4.009 |
| CsWRKY20 | TRINITY_DN5707_c0_g1.p1 | 253 | 27 820.48 | 8.97 | 66.68 | -0.733 | 4.411 |
| CsWRKY21 | TRINITY_DN238_c2_g1.p1 | 520 | 55 805.56 | 5.75 | 60.35 | -0.569 | 4.501 |
| CsWRKY22 | TRINITY_DN12424_c0_g1.p1 | 352 | 39 065.32 | 9.84 | 66.22 | -0.782 | 4.599 |
| CsWRKY23 | TRINITY_DN4346_c0_g1.p1 | 309 | 33 839.20 | 7.73 | 52.72 | -0.891 | 4.207 |
| CsWRKY24 | TRINITY_DN8931_c0_g1.p1 | 502 | 54 633.32 | 6.44 | 57.45 | -0.878 | 3.872 |
| CsWRKY25 | TRINITY_DN32124_c3_g1.p1 | 333 | 35 836.41 | 7.66 | 64.23 | -0.535 | 3.024 |
| CsWRKY26 | TRINITY_DN26434_c0_g1.p1 | 352 | 38 775.68 | 6.14 | 50.99 | -0.985 | 4.718 |
| CsWRKY27 | TRINITY_DN714_c2_g1.p1 | 298 | 32 980.16 | 6.20 | 48.12 | -0.912 | 3.346 |
| CsWRKY28 | TRINITY_DN3255_c1_g1.p1 | 722 | 78 422.37 | 5.95 | 48.49 | -0.854 | 3.944 |
| CsWRKY29 | TRINITY_DN4758_c0_g1.p1 | 284 | 30 970.33 | 5.31 | 60.11 | -0.711 | 3.870 |
| CsWRKY30 | TRINITY_DN82013_c0_g1.p1 | 204 | 23 014.52 | 7.04 | 47.79 | -0.749 | 3.481 |
| CsWRKY31 | TRINITY_DN54950_c0_g1.p1 | 121 | 13 931.73 | 9.25 | 62.07 | -1.107 | 2.837 |
| CsWRKY32 | TRINITY_DN44830_c0_g1.p1 | 99 | 11 695.20 | 9.23 | 58.08 | -0.890 | 2.449 |
| CsWRKY33 | TRINITY_DN4854_c0_g1.p1 | 318 | 35 704.86 | 7.11 | 55.50 | -0.876 | 4.841 |
| CsWRKY34 | TRINITY_DN28623_c0_g1.p1 | 234 | 26 682.65 | 9.28 | 51.62 | -0.981 | 4.762 |
| CsWRKY35 | TRINITY_DN15_c2_g1.p1 | 458 | 51 061.95 | 5.85 | 47.53 | -0.967 | 4.421 |
| CsWRKY36 | TRINITY_DN7274_c0_g1.p1 | 394 | 43 242.46 | 8.51 | 57.13 | -0.808 | 4.270 |
| CsWRKY37 | TRINITY_DN9415_c1_g1.p1 | 195 | 22 127.53 | 7.64 | 53.49 | -0.791 | 3.354 |
| CsWRKY38 | TRINITY_DN87366_c0_g1.p1 | 247 | 28 132.69 | 5.20 | 75.75 | -0.517 | 2.689 |
| CsWRKY39 | TRINITY_DN50547_c0_g2.p1 | 115 | 13 177.86 | 9.23 | 58.52 | -0.937 | 2.686 |
| CsWRKY40 | TRINITY_DN5168_c0_g1.p1 | 293 | 32 279.70 | 5.58 | 70.31 | -0.705 | 3.545 |
| CsWRKY41 | TRINITY_DN3189_c2_g1.p1 | 227 | 25 865.76 | 8.86 | 78.06 | -0.437 | 1.947 |
| 蛋白质名称 Protein name | α螺旋 Alpha-helix | 3_10螺旋 3_10 helix | 延伸链 Extended strand | 氢键转角 Turn | 弯曲 Bend | 无规卷曲 Random coil | β折叠 Beta-sheet | π-螺旋 π-helix |
|---|---|---|---|---|---|---|---|---|
| CsWRKY1 | — | — | 27.31 | 8.81 | 6.17 | 57.71 | — | — |
| CsWRKY2 | 23.84 | — | 10.41 | 2.74 | 2.74 | 60.27 | — | — |
| CsWRKY3 | 15.36 | — | 13.62 | 4.35 | 3.19 | 63.48 | — | — |
| CsWRKY4 | 12.12 | 0.23 | 15.15 | 5.13 | 4.20 | 63.17 | — | — |
| CsWRKY5 | 31.28 | — | 15.42 | 3.52 | 3.52 | 46.26 | — | — |
| CsWRKY6 | 27.76 | 0.38 | 5.99 | 4.09 | 1.33 | 60.46 | — | — |
| CsWRKY7 | 35.94 | 0.94 | 11.25 | 3.75 | 3.44 | 44.69 | — | — |
| CsWRKY8 | 3.11 | — | 17.46 | 3.11 | 5.02 | 71.05 | 0.24 | — |
| CsWRKY9 | 32.67 | — | 9.82 | 2.00 | 2.20 | 53.31 | — | — |
| CsWRKY10 | 27.87 | 1.05 | 10.10 | 4.18 | 3.48 | 53.31 | — | — |
| CsWRKY11 | 5.51 | — | 24.41 | 6.30 | 9.45 | 54.33 | — | — |
| CsWRKY12 | 13.95 | — | 9.20 | 3.56 | 4.15 | 69.14 | — | — |
| CsWRKY13 | 4.84 | 1.61 | 17.74 | 5.91 | 6.45 | 63.44 | — | — |
| CsWRKY14 | 11.29 | 2.15 | 16.13 | 3.76 | 5.38 | 61.29 | — | — |
| CsWRKY15 | 10.59 | 0.42 | 19.07 | 5.08 | 3.39 | 61.44 | — | — |
| CsWRKY16 | 25.11 | — | 8.15 | 2.58 | 3.00 | 61.16 | — | — |
| CsWRKY17 | 3.89 | — | 14.44 | 4.44 | 6.11 | 71.11 | — | — |
| CsWRKY18 | 15.77 | — | 18.47 | 3.15 | 4.50 | 58.11 | — | — |
| CsWRKY19 | 17.67 | — | 15.95 | 5.17 | 5.17 | 56.03 | — | — |
| CsWRKY20 | 18.58 | 1.19 | 15.02 | 3.95 | 3.56 | 57.71 | — | — |
| CsWRKY21 | 18.27 | 0.38 | 8.08 | 4.23 | 3.27 | 65.77 | — | — |
| CsWRKY22 | 24.43 | — | 11.36 | 3.98 | 4.26 | 55.97 | — | — |
| CsWRKY23 | 4.53 | 0.65 | 8.74 | 8.09 | 3.56 | 74.43 | — | — |
| CsWRKY24 | 5.98 | — | 16.53 | 2.99 | 3.78 | 70.72 | — | — |
| CsWRKY25 | 15.92 | 1.50 | 12.91 | 6.91 | 2.40 | 60.36 | — | — |
| CsWRKY26 | 5.40 | 0.57 | 13.35 | 3.98 | 4.26 | 72.16 | 0.28 | — |
| CsWRKY27 | 10.07 | — | 12.42 | 6.04 | 2.68 | 68.79 | — | — |
| CsWRKY28 | 9.00 | 0.28 | 9.56 | 4.43 | 3.19 | 73.55 | — | — |
| CsWRKY29 | 7.39 | 0.70 | 10.92 | 5.99 | 3.52 | 71.48 | — | — |
| CsWRKY30 | 7.35 | — | 17.16 | 4.41 | 3.43 | 67.65 | — | — |
| CsWRKY31 | 18.18 | — | 21.49 | 8.26 | 5.79 | 46.28 | — | — |
| CsWRKY32 | 11.11 | — | 33.33 | 7.07 | 14.14 | 34.34 | — | — |
| CsWRKY33 | 14.15 | 0.31 | 14.47 | 2.83 | 3.14 | 65.09 | — | — |
| CsWRKY34 | 4.27 | — | 14.10 | 2.56 | 5.98 | 73.08 | — | — |
| CsWRKY35 | 6.33 | — | 17.03 | 4.80 | 4.37 | 67.47 | — | — |
| CsWRKY36 | 5.33 | — | 18.53 | 4.06 | 4.82 | 67.26 | — | — |
| CsWRKY37 | 3.08 | — | 23.59 | 3.59 | 5.64 | 64.10 | — | — |
| CsWRKY38 | 23.89 | — | 14.57 | 4.86 | 4.45 | 52.23 | — | — |
| CsWRKY39 | 10.43 | — | 26.96 | 3.48 | 7.83 | 51.30 | — | — |
| CsWRKY40 | 23.89 | — | 10.92 | 5.12 | 2.73 | 57.34 | — | — |
| CsWRKY41 | 19.38 | — | 11.45 | 6.61 | 2.64 | 59.91 | — | — |
表3 CsWRKY家族蛋白质的二级结构占比
Table 3 Proportion of secondary structure of CsWRKY family proteins %
| 蛋白质名称 Protein name | α螺旋 Alpha-helix | 3_10螺旋 3_10 helix | 延伸链 Extended strand | 氢键转角 Turn | 弯曲 Bend | 无规卷曲 Random coil | β折叠 Beta-sheet | π-螺旋 π-helix |
|---|---|---|---|---|---|---|---|---|
| CsWRKY1 | — | — | 27.31 | 8.81 | 6.17 | 57.71 | — | — |
| CsWRKY2 | 23.84 | — | 10.41 | 2.74 | 2.74 | 60.27 | — | — |
| CsWRKY3 | 15.36 | — | 13.62 | 4.35 | 3.19 | 63.48 | — | — |
| CsWRKY4 | 12.12 | 0.23 | 15.15 | 5.13 | 4.20 | 63.17 | — | — |
| CsWRKY5 | 31.28 | — | 15.42 | 3.52 | 3.52 | 46.26 | — | — |
| CsWRKY6 | 27.76 | 0.38 | 5.99 | 4.09 | 1.33 | 60.46 | — | — |
| CsWRKY7 | 35.94 | 0.94 | 11.25 | 3.75 | 3.44 | 44.69 | — | — |
| CsWRKY8 | 3.11 | — | 17.46 | 3.11 | 5.02 | 71.05 | 0.24 | — |
| CsWRKY9 | 32.67 | — | 9.82 | 2.00 | 2.20 | 53.31 | — | — |
| CsWRKY10 | 27.87 | 1.05 | 10.10 | 4.18 | 3.48 | 53.31 | — | — |
| CsWRKY11 | 5.51 | — | 24.41 | 6.30 | 9.45 | 54.33 | — | — |
| CsWRKY12 | 13.95 | — | 9.20 | 3.56 | 4.15 | 69.14 | — | — |
| CsWRKY13 | 4.84 | 1.61 | 17.74 | 5.91 | 6.45 | 63.44 | — | — |
| CsWRKY14 | 11.29 | 2.15 | 16.13 | 3.76 | 5.38 | 61.29 | — | — |
| CsWRKY15 | 10.59 | 0.42 | 19.07 | 5.08 | 3.39 | 61.44 | — | — |
| CsWRKY16 | 25.11 | — | 8.15 | 2.58 | 3.00 | 61.16 | — | — |
| CsWRKY17 | 3.89 | — | 14.44 | 4.44 | 6.11 | 71.11 | — | — |
| CsWRKY18 | 15.77 | — | 18.47 | 3.15 | 4.50 | 58.11 | — | — |
| CsWRKY19 | 17.67 | — | 15.95 | 5.17 | 5.17 | 56.03 | — | — |
| CsWRKY20 | 18.58 | 1.19 | 15.02 | 3.95 | 3.56 | 57.71 | — | — |
| CsWRKY21 | 18.27 | 0.38 | 8.08 | 4.23 | 3.27 | 65.77 | — | — |
| CsWRKY22 | 24.43 | — | 11.36 | 3.98 | 4.26 | 55.97 | — | — |
| CsWRKY23 | 4.53 | 0.65 | 8.74 | 8.09 | 3.56 | 74.43 | — | — |
| CsWRKY24 | 5.98 | — | 16.53 | 2.99 | 3.78 | 70.72 | — | — |
| CsWRKY25 | 15.92 | 1.50 | 12.91 | 6.91 | 2.40 | 60.36 | — | — |
| CsWRKY26 | 5.40 | 0.57 | 13.35 | 3.98 | 4.26 | 72.16 | 0.28 | — |
| CsWRKY27 | 10.07 | — | 12.42 | 6.04 | 2.68 | 68.79 | — | — |
| CsWRKY28 | 9.00 | 0.28 | 9.56 | 4.43 | 3.19 | 73.55 | — | — |
| CsWRKY29 | 7.39 | 0.70 | 10.92 | 5.99 | 3.52 | 71.48 | — | — |
| CsWRKY30 | 7.35 | — | 17.16 | 4.41 | 3.43 | 67.65 | — | — |
| CsWRKY31 | 18.18 | — | 21.49 | 8.26 | 5.79 | 46.28 | — | — |
| CsWRKY32 | 11.11 | — | 33.33 | 7.07 | 14.14 | 34.34 | — | — |
| CsWRKY33 | 14.15 | 0.31 | 14.47 | 2.83 | 3.14 | 65.09 | — | — |
| CsWRKY34 | 4.27 | — | 14.10 | 2.56 | 5.98 | 73.08 | — | — |
| CsWRKY35 | 6.33 | — | 17.03 | 4.80 | 4.37 | 67.47 | — | — |
| CsWRKY36 | 5.33 | — | 18.53 | 4.06 | 4.82 | 67.26 | — | — |
| CsWRKY37 | 3.08 | — | 23.59 | 3.59 | 5.64 | 64.10 | — | — |
| CsWRKY38 | 23.89 | — | 14.57 | 4.86 | 4.45 | 52.23 | — | — |
| CsWRKY39 | 10.43 | — | 26.96 | 3.48 | 7.83 | 51.30 | — | — |
| CsWRKY40 | 23.89 | — | 10.92 | 5.12 | 2.73 | 57.34 | — | — |
| CsWRKY41 | 19.38 | — | 11.45 | 6.61 | 2.64 | 59.91 | — | — |
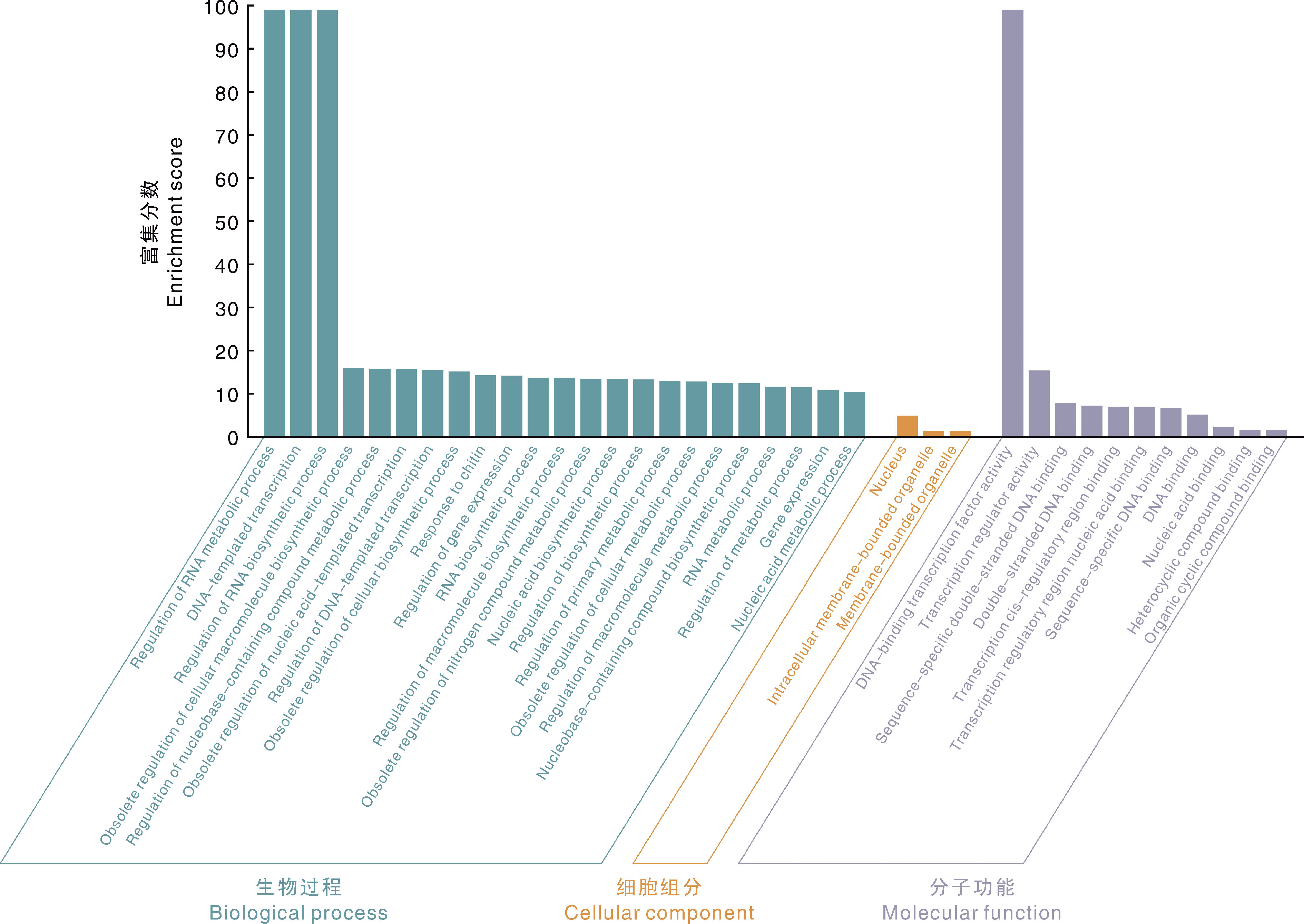
图5 CsWRKY家族GO富集分析 Regulation of RNA metabolic process,RNA代谢过程调控;DNA-templated transcription,DNA模板转录;Regulation of RNA biosynthetic process,RNA生物合成过程调控;Obsolete regulation of cellular macromolecule biosynthetic process,细胞大分子生物合成过程的过时调控;Regulation of nucleobase-containing compound metabolic process,含核碱基化合物代谢过程的调控;Obsolete regulation of nucleic acid-templated transcription,核酸模板转录的过时调控;Regulation of DNA-templated transcription,DNA模板转录的调控;Obsolete regulation of cellular biosynthetic process,细胞生物合成过程的过时调控;Response to chitin,对几丁质的反应;Regulation of gene expression,基因表达调控;RNA biosynthetic process,RNA生物合成过程;Regulation of macromolecule biosynthetic process,大分子生物合成过程的调控;Obsolete regulation of nitrogen compound metabolic process,氮化合物代谢过程的过时调控;Nucleic acid biosynthetic process,核酸生物合成过程;Regulation of biosynthetic process,生物合成过程的调控;Regulation of primary metabolic process,初级代谢过程的调节;Obsolete regulation of cellular metabolic process,细胞代谢过程的过时调节;Regulation of macromolecule metabolic process,大分子代谢过程的调控;Nucleobase-containing compound biosynthetic process,含核碱基化合物生物合成过程;RNA metabolic process,RNA代谢过程;Regulation of metabolic process,代谢过程的调节;gene expression,基因表达;Nucleic acid metabolic process,核酸代谢过程;Nucleus,细胞核;Intracellular membrane-bounded organelle,细胞内的膜结构细胞器;Membrane-bounded organelle,膜结构细胞器;DNA-binding transcription factor activity,DNA结合转录因子活性;Transcription regulator activity,转录调节因子活性;Sequence-specific double-stranded DNA binding,序列特异性双链DNA结合;Double-stranded DNA binding,双链DNA结合;Transcription cis-regulatory region binding,转录顺式调控区域结合;Transcription regulatory region nucleic acid binding,转录调控区域核酸结合;Sequence-specific DNA binding,序列特异性DNA结合;DNA binding,DNA结合;Nucleic acid binding,核酸结合;Heterocyclic compound binding,杂环化合物结合;Organic cyclic compound binding,有机环状化合物结合。
Fig.5 GO enrichment analysis of CsWRKY family
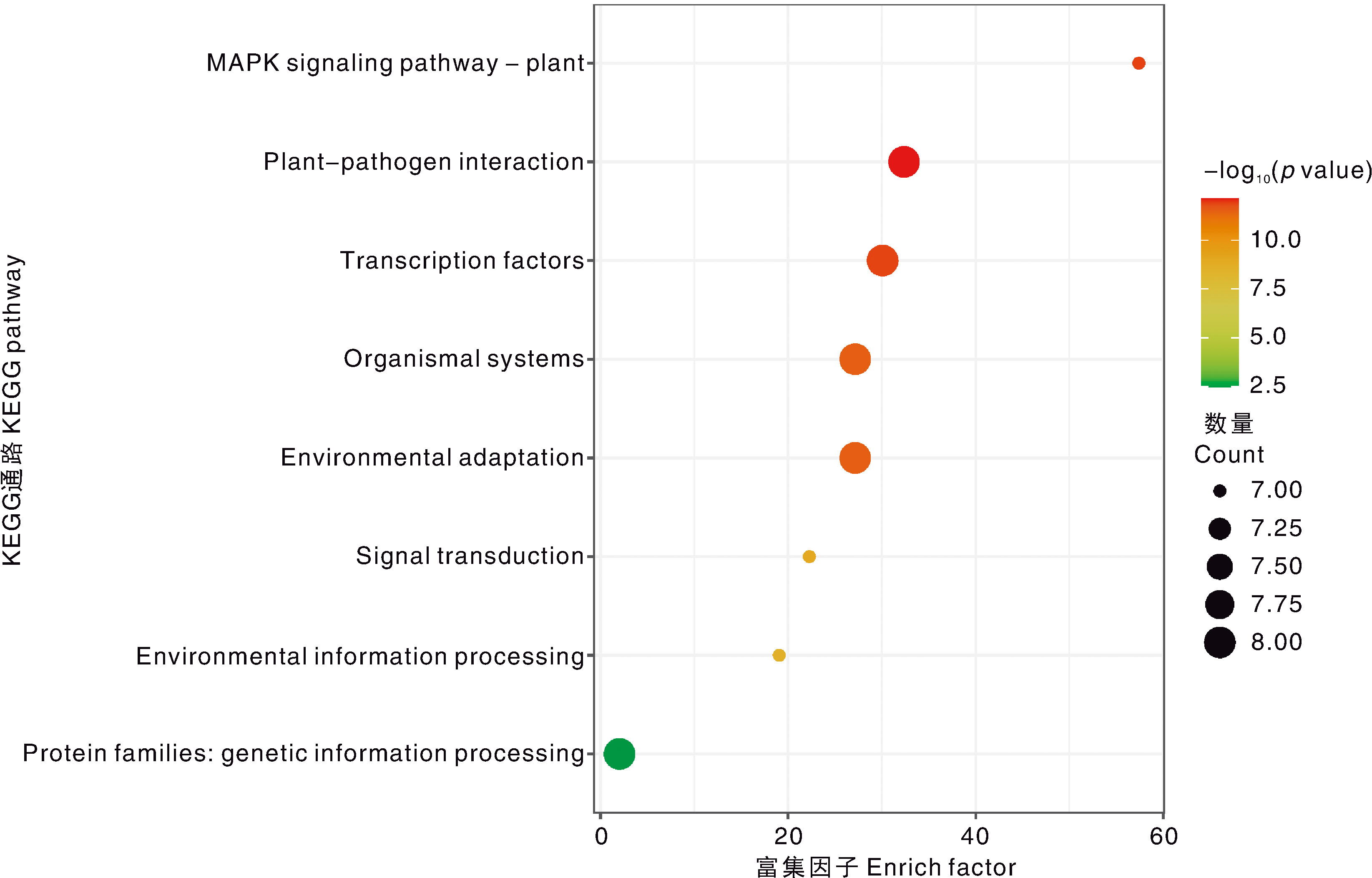
图6 CsWRKY家族的KEGG通路 MAPK signaling pathway-plant,植物MAPK信号通路;Plant-pathogen interaction,植物-病原互作;Transcription factors,转录因子;Organismal systems,生物系统;Environmental adaptation,环境适应;Signal transduction,信号传导;Environmental information processing,环境信息处理;Protein families: genetic information processing,蛋白质家族:遗传信息处理。
Fig.6 KEGG pathway of the CsWRKY family
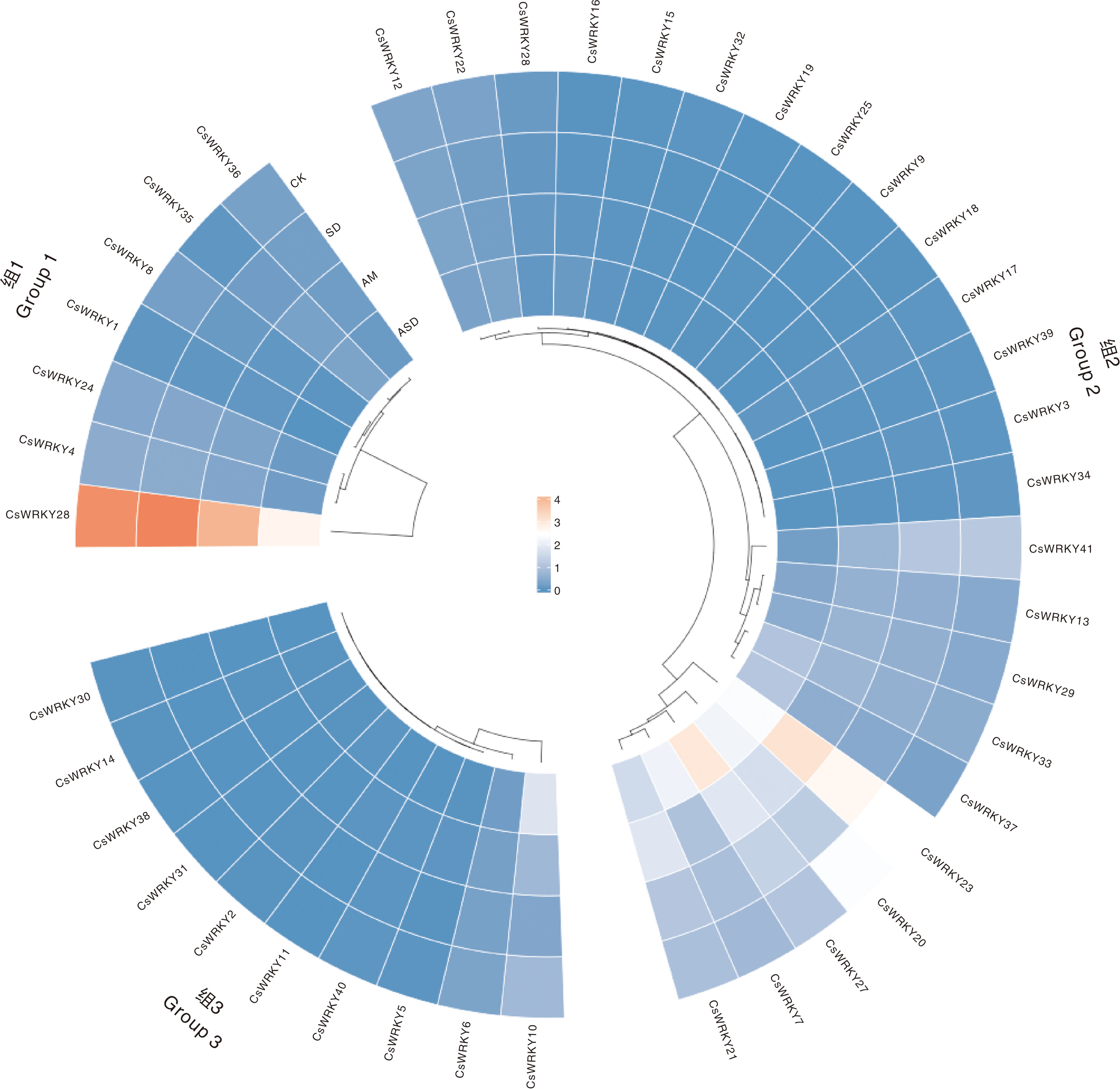
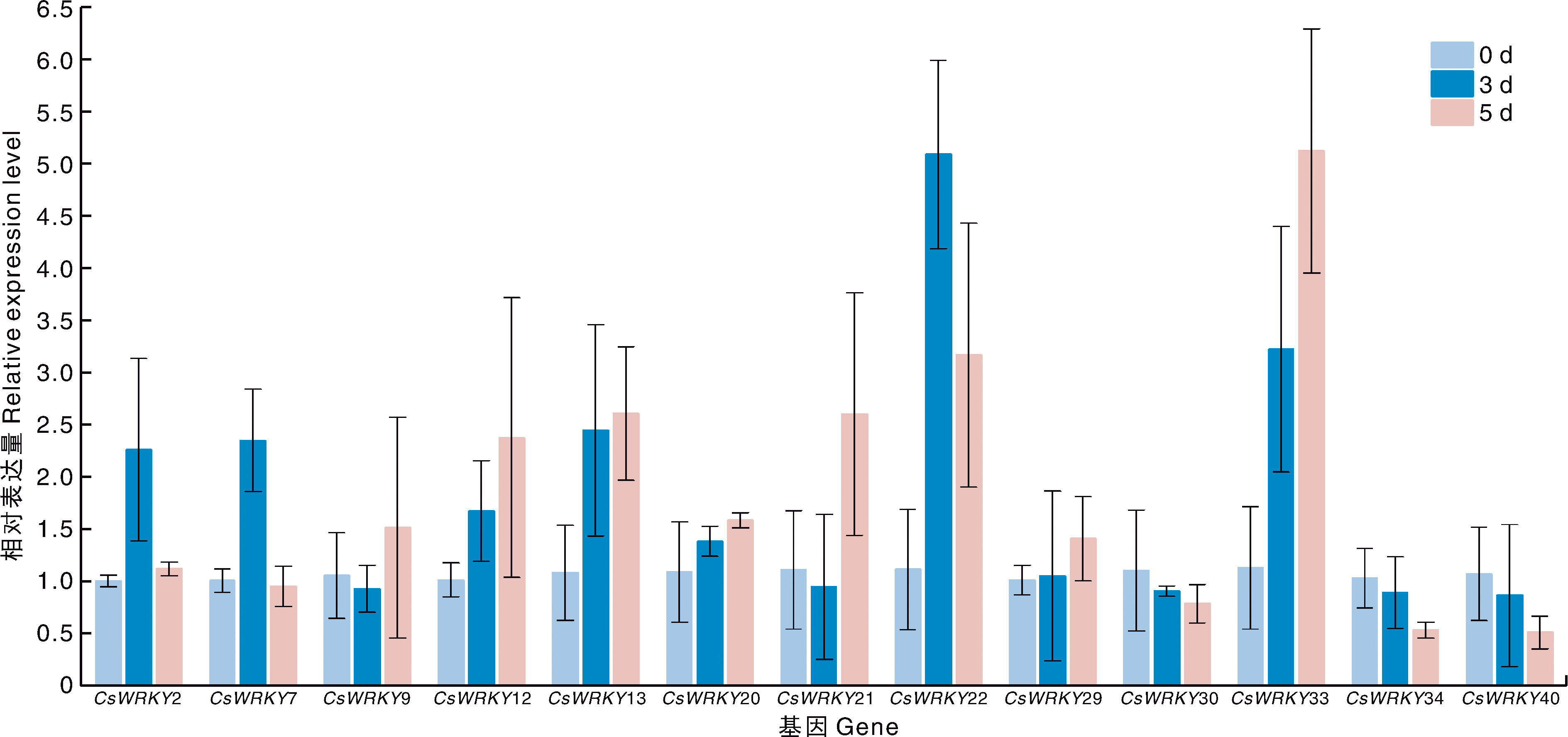
图7 CsWRKY基因在不同处理下的表达模式 CK,对照组;SD,干旱处理;AM,接种AM真菌处理;ASD,接种AM真菌同时干旱处理。图8同。
Fig.7 Expression patterns of CsWRKY genes under different treatments CK, Control group; SD, Drought treatment; AM, AM treatment; ASD, AM and drought treatment. The same as in Fig. 8.
| AtWRKY | CsWRKY候选基因 Candidate CsWRKY gene | BLAST位分数 BLAST bit score | AtWRKY | CsWRKY候选基因 Candidate CsWRKY gene | BLAST位分数 BLAST bit score |
|---|---|---|---|---|---|
| AtWRKY11[ | CsWRKY22 CsWRKY12 CsWRKY29 | 186.0 181.0 106.0 | AtWRKY40[ | CsWRKY7 CsWRKY20 CsWRKY9 | 201.0 176.0 103.0 |
| AtWRKY17[ | CsWRKY22 CsWRKY12 CsWRKY29 | 181.0 178.0 103.0 | AtWRKY46[ | CsWRKY40 CsWRKY2 CsWRKY30 | 120.0 92.4 89.0 |
| AtWRKY18[ | CsWRKY7 CsWRKY20 CsWRKY21 | 209.0 171.0 115.0 | AtWRKY57[ | CsWRKY13 CsWRKY33 CsWRKY34 | 154.0 154.0 129.0 |
| AtWRKY28[ | CsWRKY33 CsWRKY13 CsWRKY34 | 182.0 132.0 118.0 | AtWRKY60[ | CsWRKY7 CsWRKY20 CsWRKY9 | 193.0 165.0 107.0 |
| AtWRKY30[ | CsWRKY2 CsWRKY40 CsWRKY30 | 96.7 88.6 81.3 |
表4 耐干旱胁迫的候选CsWRKY基因
Table 4 Candidate CsWRKY genes for drought tolerance
| AtWRKY | CsWRKY候选基因 Candidate CsWRKY gene | BLAST位分数 BLAST bit score | AtWRKY | CsWRKY候选基因 Candidate CsWRKY gene | BLAST位分数 BLAST bit score |
|---|---|---|---|---|---|
| AtWRKY11[ | CsWRKY22 CsWRKY12 CsWRKY29 | 186.0 181.0 106.0 | AtWRKY40[ | CsWRKY7 CsWRKY20 CsWRKY9 | 201.0 176.0 103.0 |
| AtWRKY17[ | CsWRKY22 CsWRKY12 CsWRKY29 | 181.0 178.0 103.0 | AtWRKY46[ | CsWRKY40 CsWRKY2 CsWRKY30 | 120.0 92.4 89.0 |
| AtWRKY18[ | CsWRKY7 CsWRKY20 CsWRKY21 | 209.0 171.0 115.0 | AtWRKY57[ | CsWRKY13 CsWRKY33 CsWRKY34 | 154.0 154.0 129.0 |
| AtWRKY28[ | CsWRKY33 CsWRKY13 CsWRKY34 | 182.0 132.0 118.0 | AtWRKY60[ | CsWRKY7 CsWRKY20 CsWRKY9 | 193.0 165.0 107.0 |
| AtWRKY30[ | CsWRKY2 CsWRKY40 CsWRKY30 | 96.7 88.6 81.3 |
| [1] | HUANG J P, YU H P, DAI A G, et al. Drylands face potential threat under 2 ℃ global warming target[J]. Nature Climate Change, 2017, 7(6): 417-422. |
| [2] | 马住国, 符淙斌, 杨庆, 等. 关于我国北方干旱化及其转折性变化[J]. 大气科学, 2018, 42(4): 951-961. |
| MA Z G, FU C B, YANG Q, et al. Drying trend in Northern China and its shift during 1951-2016[J]. Chinese Journal of Atmospheric Sciences, 2018, 42(4): 951-961. | |
| [3] | 陈兰兰, 王丽, 吴亚娟, 等. 植物响应干旱胁迫的分子和微生态机制[J/OL]. 分子植物育种, 2023: 1-15. [2024-12-10]. https://kns.cnki.net/kcms/detail/46.1068.S.20230406.1634.006.html. |
| CHEN L L, WANG L, WU Y J, et al. Molecular and microecological mechanisms of plant responses to drought stress[J/OL]. Molecular Plant Breeding, 2023: 1-15. [2024-12-10]. https://kns.cnki.net/kcms/detail/46.1068.S.20230406.1634.006.html. (in Chinese with English abstract) | |
| [4] | 刘晨, 徐浩博, 斯钰阳, 等. 基于转录组学的植物响应盐胁迫调控机制研究进展[J]. 浙江农业学报, 2022, 34(4): 870-878. |
| LIU C, XU H B, SI Y Y, et al. Research progress on regulation mechanism of plant response to salt stress based on transcriptomics[J]. Acta Agriculturae Zhejiangensis, 2022, 34(4): 870-878. (in Chinese with English abstract) | |
| [5] | 陈丽飞, 李嘉峻, 刘云怡慧, 等. 干旱胁迫下大苞萱草WRKY基因家族成员鉴定及生物信息学分析[J]. 河南农业科学, 2023, 52(2): 113-123. |
| CHEN L F, LI J J, LIU Y, et al. Identification and bioinformatics analysis of WRKY gene family members in Hemerocallis middendorffii under drought stress[J]. Journal of Henan Agricultural Sciences, 2023, 52(2): 113-123. (in Chinese with English abstract) | |
| [6] | WU K L. The WRKY family of transcription factors in rice and Arabidopsis and their origins[J]. DNA Research, 2005, 12(1): 9-26. |
| [7] | EULGEM T, RUSHTON P J, ROBATZEK S, et al. The WRKY superfamily of plant transcription factors[J]. Trends in Plant Science, 2000, 5(5): 199-206. |
| [8] | BABITHA K C, RAMU S V, PRUTHVI V, et al. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis[J]. Transgenic Research, 2013, 22(2): 327-341. |
| [9] | 张亚萱, 李洪臣, 李丽华, 等. 烟草转录因子NtWRKY65在响应干旱胁迫中的功能研究[J]. 江苏农业科学, 2025, 53(4): 225-233. |
| ZHANG Y X, LI H C, LI L H, et al. Functional analysis of transcription factor NtWRKY65 in tobacco under drought stress[J]. Jiangsu Agricultural Sciences, 2025, 53(4): 225-233. (in Chinese with English abstract) | |
| [10] | 宋娟, 吴祝华, 王靖涵, 等. 丛枝菌根真菌对NaCl胁迫和非胁迫下拟南芥相关指标的影响[J]. 植物资源与环境学报, 2024, 33(6): 113-116. |
| SONG J, WU Z H, WANG J H, et al. Effects of arbuscular mycorrhizal fungi on relative indexes of Arabidopsis thaliana under NaCl stress and non-stress[J]. Journal of Plant Resources and Environment, 2024, 33(6): 113-116. (in Chinese with English abstract) | |
| [11] | QUIROGA G, ERICE G, AROCA R, et al. Elucidating the possible involvement of maize aquaporins and arbuscular mycorrhizal symbiosis in the plant ammonium and urea transport under drought stress conditions[J]. Plants, 2020, 9(2): 148. |
| [12] | SHANG P P, ZHENG R C, LI Y D, et al. Effect of AM fungi on the growth and powdery mildew development of Astragalus sinicus L. under water stress[J]. Plant Physiology and Biochemistry, 2025, 219: 109422. |
| [13] | 钟陈佺. 崖棕抗细菌化学活性成分研究[D]. 杨凌: 西北农林科技大学, 2019. |
| ZHONG C Q. Studies on the anti-bacterial constituents of Carex siderosticta Hance[D]. Yangling: Northwest A & F University, 2019. (in Chinese with English abstract) | |
| [14] | 王艺璇. 冻融循环对宽叶薹草根际土壤与植株生理特性的影响[D]. 哈尔滨: 东北林业大学, 2023. |
| WANG Y X. Effects of freeze-thaw cycles on the physiological characteristics of Carex siderosticta Hance inter-rooted soils and plant[D]. Harbin: Northeast Forestry University, 2023. (in Chinese with English abstract) | |
| [15] | MISTRY J, CHUGURANSKY S, WILLIAMS L, et al. Pfam: the protein families database in 2021[J]. Nucleic Acids Research, 2021, 49(D1): D412-D419. |
| [16] | TAMURA K, PETERSON D, PETERSON N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods[J]. Molecular Biology and Evolution, 2011, 28(10): 2731-2739. |
| [17] | CHEN C J, WU Y, LI J W, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining[J]. Molecular Plant, 2023, 16(11): 1733-1742. |
| [18] | 梁锦, 刘海婷, 钟荣, 等. 萱草不同器官实时荧光定量PCR内参基因的筛选[J]. 植物生理学报, 2020, 56(9): 1891-1898. |
| LIANG J, LIU H T, ZHONG R, et al. Screening of reference genes for quantitative real-time PCR in different organs of Hemerocallis fulva[J]. Plant Physiology Journal, 2020, 56(9): 1891-1898. (in Chinese with English abstract) | |
| [19] | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 - Δ Δ C T method[J]. Methods, 2001, 25(4): 402-408. |
| [20] | ALI M A, AZEEM F, NAWAZ M A, et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis[J]. Journal of Plant Physiology, 2018, 226: 12-21. |
| [21] | CHEN H, LAI Z B, SHI J W, et al. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress[J]. BMC Plant Biology, 2010, 10: 281. |
| [22] | SCARPECI T E, ZANOR M I, MUELLER-ROEBER B, et al. Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana[J]. Plant Molecular Biology, 2013, 83(3): 265-277. |
| [23] | 车永梅, 孙艳君, 卢松冲, 等. AtWRKY40参与拟南芥干旱胁迫响应过程[J]. 植物生理学报, 2018, 54(3): 456-464. |
| CHE Y M, SUN Y J, LU S C, et al. AtWRKY40 functions in drought stress response in Arabidopsis thaliana[J]. Plant Physiology Journal, 2018, 54(3): 456-464. (in Chinese with English abstract) | |
| [24] | DING Z J, YAN J Y, XU X Y, et al. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis[J]. The Plant Journal, 2014, 79(1): 13-27. |
| [25] | JIANG Y J, LIANG G, YU D Q. Activated expression of WRKY57 confers drought tolerance in Arabidopsis[J]. Molecular Plant, 2012, 5(6): 1375-1388. |
| [26] | EULGEM T, SOMSSICH I E. Networks of WRKY transcription factors in defense signaling[J]. Current Opinion in Plant Biology, 2007, 10(4): 366-371. |
| [27] | ROSS C A, LIU Y, SHEN Q J. The WRKY gene family in rice (Oryza sativa)[J]. Journal of Integrative Plant Biology, 2007, 49(6): 827-842. |
| [28] | BENCKE-MALATO M, CABREIRA C, WIEBKE-STROHM B, et al. Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection[J]. BMC Plant Biology, 2014, 14: 236. |
| [29] | 宋辉, 南志标. 蒺藜苜蓿全基因组中WRKY转录因子的鉴定与分析[J]. 遗传, 2014, 36(2): 152-168. |
| SONG H, NAN Z B. Genome-wide identification and analysis of WRKY transcription factors in Medicago truncatula[J]. Hereditas, 2014, 36(2): 152-168. (in Chinese with English abstract) | |
| [30] | WANG H P, CHEN W Q, XU Z Y, et al. Functions of WRKYs in plant growth and development[J]. Trends in Plant Science, 2023, 28(6): 630-645. |
| [31] | ZHOU H, ZHU W, WANG X C, et al. A missense mutation in WRKY32 converts its function from a positive regulator to a repressor of photomorphogenesis[J]. New Phytologist, 2022, 235(1): 111-125. |
| [32] | YIN M, SONG N, CHEN S Y, et al. NaKTI2, a Kunitz trypsin inhibitor transcriptionally regulated by NaWRKY3 and NaWRKY6, is required for herbivore resistance in Nicotiana attenuata[J]. Plant Cell Reports, 2021, 40(1): 97-109. |
| [33] | WANG L J, GUO D Z, ZHAO G D, et al. Group IIc WRKY transcription factors regulate cotton resistance to Fusarium oxysporum by promoting GhMKK2-mediated flavonoid biosynthesis[J]. New Phytologist, 2022, 236(1): 249-265. |
| [34] | LI W X, PANG S Y, LU Z G, et al. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants[J]. Plants, 2020, 9(11): 1515. |
| [35] | FAZAL F M, CHANG H Y. Subcellular spatial transcriptomes: emerging frontier for understanding gene regulation[J]. Cold Spring Harbor Symposia on Quantitative Biology, 2019, 84: 31-45. |
| [36] | 刘晨, 曹小汉, 殷丹丹, 等. MAPK信号通路调控植物响应非生物胁迫的研究进展[J]. 安徽农业科学, 2022, 50(18): 9-16. |
| LIU C, CAO X H, YIN D D, et al. Research progress of MAPK signaling pathway in regulating plants response to abiotic stress[J]. Journal of Anhui Agricultural Sciences, 2022, 50(18): 9-16. (in Chinese with English abstract) | |
| [37] | ZHENG Z Y, MOSHER S L, FAN B F, et al. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae[J]. BMC Plant Biology, 2007, 7: 2. |
| [38] | 王怡, 于月华, 万会娜, 等. GhWRKY44在干旱胁迫下的功能鉴定[J]. 棉花学报, 2024, 36(4): 275-284. |
| WANG Y, YU Y H, WAN H N, et al. Functional identification of GhWRKY44 under drought stress[J]. Cotton Science, 2024, 36(4): 275-284. (in Chinese with English abstract) | |
| [39] | LEE H, CHA J, CHOI C, et al. Rice WRKY11 plays a role in pathogen defense and drought tolerance[J]. Rice, 2018, 11(1): 5. |
| [40] | EL-ESAWI M A, AL-GHAMDI A A, ALI H M, et al. Overexpression of AtWRKY30 transcription factor enhances heat and drought stress tolerance in wheat (Triticum aestivum L.)[J]. Genes, 2019, 10(2): 163. |
| [41] | REN X Z, CHEN Z Z, LIU Y, et al. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis[J]. Plant Journal, 2010, 63(3): 417-429. |
| [42] | QIAO Z, LI C L, ZHANG W. WRKY1 regulates stomatal movement in drought-stressed Arabidopsis thaliana[J]. Plant Molecular Biology, 2016, 91(1): 53-65. |
| [43] | WANG X J, DU B J, LIU M, et al. Arabidopsis transcription factor WRKY33 is involved in drought by directly regulating the expression of CesA8[J]. American Journal of Plant Sciences, 2013, 4(6): 21-27. |
| [1] | 李宇静, 黄倩茹, 张爱冬, 吴雪霞, 朱栋幸, 肖凯. 茄子SmMYB13基因在干旱胁迫响应中的功能[J]. 浙江农业学报, 2025, 37(8): 1666-1679. |
| [2] | 师阳阳, 吕丽霞, 脱登峰. 低温弱光胁迫下AMF和PGPR对紫罗兰生长及营养吸收的影响[J]. 浙江农业学报, 2025, 37(8): 1694-1705. |
| [3] | 狄延翠, 嵇泽琳, 王媛媛, 娄世浩, 张涛, 国志信, 申顺善, 朴凤植, 杜南山, 董晓星, 董韩. 番茄SlMYB52基因鉴定、亚细胞定位及表达分析[J]. 浙江农业学报, 2025, 37(4): 808-819. |
| [4] | 张美莹, 莫倩, 齐秀双, 佟宁宁, 孔凡, 刘政安, 吕长平, 彭丽平. 牡丹PoLPAT2基因的克隆及表达分析[J]. 浙江农业学报, 2025, 37(2): 321-328. |
| [5] | 任元龙, 马蓉, 王晓卓, 张雪艳. 叶面喷施褪黑素对甘蓝幼苗干旱胁迫的缓解作用[J]. 浙江农业学报, 2025, 37(2): 338-348. |
| [6] | 闵江艳, 唐卓磊, 杨雪, 黄小燕, 黄凯丰, 何佩云. 不同干旱-复水模式对苦荞生长与产量的影响[J]. 浙江农业学报, 2024, 36(9): 2000-2009. |
| [7] | 蒋文骏, 舒红锁, 陈正满, 任典挺, 杨党, 田荣江, 杜照奎. 秋茄KoWRKY43基因克隆、表达与生物信息学分析[J]. 浙江农业学报, 2024, 36(8): 1832-1843. |
| [8] | 杨明凤, 吉春容, 刘勇, 白书军, 陈雪, 刘爱琳. 花铃期持续干旱胁迫对棉花生长与土壤干旱阈值的影响[J]. 浙江农业学报, 2024, 36(4): 738-747. |
| [9] | 李亚萍, 金福来, 黄宗贵, 张涛, 段晓婧, 姜武, 陶正明, 陈家栋. 铁皮石斛糖苷水解酶GH3基因家族鉴定及表达模式分析[J]. 浙江农业学报, 2024, 36(4): 790-799. |
| [10] | 张露荷, 王多锋, 张德, 张广忠, 赵通, 吕斌燕, 张洋军, 李毅. 枣树novel-miR16靶基因ZjTCP4鉴定及生物信息学分析[J]. 浙江农业学报, 2024, 36(3): 534-543. |
| [11] | 田晓明, 向光锋, 牟村, 吕浩, 马涛, 朱路, 彭静, 张敏, 何艳. 四种红豆属植物耐旱性综合评价[J]. 浙江农业学报, 2024, 36(2): 308-324. |
| [12] | 张丽, 王媛媛, 王瑞, 刘丽霞. 牦牛DRA基因克隆测序及生物信息学分析[J]. 浙江农业学报, 2023, 35(7): 1564-1570. |
| [13] | 庞雪晴, 唐诗, 曾红梅, 赵位, 王印, 罗燕, 姚学萍, 任梅渗, 任永军, 杨泽晓. 两株GI.1型和GI.2型兔出血症病毒RdRp基因的克隆与分析[J]. 浙江农业学报, 2023, 35(6): 1286-1296. |
| [14] | 张新业, 李文静, 朱姝, 孙艳香, 王聪艳, 闫训友, 周志国. 三种伞形科蔬菜作物棕榈酰基转移酶基因家族的鉴定与分析[J]. 浙江农业学报, 2023, 35(6): 1315-1327. |
| [15] | 宋雅萍, 雷召雄, 赵毅昂, 姜超, 王兴平, 罗仍卓么, 马云, 魏大为. 牛FoxO1基因CDS区克隆及其在脂肪细胞分化过程中的表达分析[J]. 浙江农业学报, 2023, 35(5): 1016-1027. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||