浙江农业学报 ›› 2026, Vol. 38 ›› Issue (1): 1-16.DOI: 10.3969/j.issn.1004-1524.20250086
大豆耐盐与耐镉胁迫共性基因的挖掘
徐燕1,2( ), 李素娟3, 陈光2, 徐盛春1,2,4, 王剑2,*(
), 李素娟3, 陈光2, 徐盛春1,2,4, 王剑2,*( )
)
- 1.浙江农林大学 现代农学院,浙江 杭州 311300
2.浙江省农业科学院 数字农业研究所,浙江 杭州 310021
3.浙江省农业科学院 农产品质量安全全国重点实验室,浙江 杭州 310021
4.湘湖实验室,浙江 杭州 311258
-
收稿日期:2025-02-07出版日期:2026-01-25发布日期:2026-02-11 -
作者简介:王剑,E-mail: wangj@zaas.ac.cn
徐燕,主要从事大豆抗性基因筛选与功能分析。E-mail:2932737880@qq.com -
通讯作者:王剑 -
基金资助:中央引导地方科技发展资金(2023ZY1016)
Identification of common genes for salt and cadmium tolerance in soybean
XU Yan1,2( ), LI Sujuan3, CHEN Guang2, XU Shengchun1,2,4, WANG Jian2,*(
), LI Sujuan3, CHEN Guang2, XU Shengchun1,2,4, WANG Jian2,*( )
)
- 1. College of Advanced Agricultural Science, Zhejiang A&F University, Hangzhou 311300, China
2. Institute of Digital Agriculture, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, China
3. State Key Laboratory for Quality and Safety of Agro-Products, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, China
4. Xianghu Laboratory, Hangzhou 311258, China
-
Received:2025-02-07Online:2026-01-25Published:2026-02-11 -
Contact:WANG Jian
摘要:
盐碱化土地偶尔会伴有重金属污染。研究植物对盐、镉复合胁迫的生长与生理响应,并挖掘其共有耐性基因和调控通路,对作物抗逆遗传改良具有重要意义。本研究以50份大豆栽培种和野生种为材料,分别设置盐胁迫(200 mmol·L-1 NaCl)、镉胁迫(0.3 mmol·L-1 CdCl2)与正常条件进行培养,测定萌发率、株高、根长、根长与株高比、地上部与地下部鲜重共6个指标,通过主成分分析筛选关键耐性指标;利用耐性最强的野生大豆种质W-3-12-90构建全长cDNA酵母表达文库,结合全长cDNA过表达(FOX)基因搜寻系统与二代测序,鉴定盐、镉共耐受相关基因。主成分分析结果显示,株高、根长、地上部鲜重和萌发率是评价大豆耐盐、耐镉能力的4项关键指标;野生大豆种质W-3-12-90在盐、镉胁迫下耐受性最强。基于该材料共鉴定出109个盐、镉共同响应基因。亚细胞定位预测显示,39个基因编码胞外蛋白,此类蛋白响应快、占比高;51个基因编码的蛋白质分布于细胞核、细胞质与细胞膜,主要参与蛋白质代谢、细胞信号转导、防御与应激反应,以及氧化还原酶活性等通路。基因表达分析表明,6个候选共耐性基因在盐、镉胁迫下均显著上调。综上,植物响应盐、镉胁迫的基因主要通过编码胞外蛋白,并借助其与质膜、细胞核及细胞质的相互作用协同调控植物的耐盐、耐镉能力。本研究为解析大豆耐盐、耐镉的分子机制提供了新思路,并为培育耐盐、耐镉新种质提供了基因资源与理论依据。
中图分类号:
引用本文
徐燕, 李素娟, 陈光, 徐盛春, 王剑. 大豆耐盐与耐镉胁迫共性基因的挖掘[J]. 浙江农业学报, 2026, 38(1): 1-16.
XU Yan, LI Sujuan, CHEN Guang, XU Shengchun, WANG Jian. Identification of common genes for salt and cadmium tolerance in soybean[J]. Acta Agriculturae Zhejiangensis, 2026, 38(1): 1-16.
| 基因Gene | 正向引物序列(5'→3') Forward primer sequence(5'→3') | 反向引物序列(5'→3') Reverse primer sequence(5'→3') |
|---|---|---|
| GmEF1b | CCACTGCTGAAGAAGATGATGATG | AAGGACAGAAGACTTGCCACTC |
| GmDehydrin | AGGAAGGAACATCGTCAGCA | TGACAAGACACTGTACGTACG |
| GmSSP | CCACCTCAGGAGTCTCAGAA | CCCGCAAAAGTTTCGTGACT |
| GmGF14 | ACGTTGGGAGAGGAATCATACA | GCATTCAACACCTTCTCCCT |
| GmPAP85 | AGCAGAAAGAGGAGGGGAAC | AGCAGACAGTTGAAGTACACA |
| GmHUP54 | ATGCCTAGGATTGACAGCGA | AGCAGAGTCAGCACCATCAT |
| GmMET2 | TCGAGAGTGCTGAAATGGGT | ACACACCCATCACAAGTCCA |
表1 耐盐、耐镉候选基因qRT-PCR分析所用的引物
Table 1 Primers for qRT-PCR analysis of candidate genes associated with salt- and cadmium-tolerance
| 基因Gene | 正向引物序列(5'→3') Forward primer sequence(5'→3') | 反向引物序列(5'→3') Reverse primer sequence(5'→3') |
|---|---|---|
| GmEF1b | CCACTGCTGAAGAAGATGATGATG | AAGGACAGAAGACTTGCCACTC |
| GmDehydrin | AGGAAGGAACATCGTCAGCA | TGACAAGACACTGTACGTACG |
| GmSSP | CCACCTCAGGAGTCTCAGAA | CCCGCAAAAGTTTCGTGACT |
| GmGF14 | ACGTTGGGAGAGGAATCATACA | GCATTCAACACCTTCTCCCT |
| GmPAP85 | AGCAGAAAGAGGAGGGGAAC | AGCAGACAGTTGAAGTACACA |
| GmHUP54 | ATGCCTAGGATTGACAGCGA | AGCAGAGTCAGCACCATCAT |
| GmMET2 | TCGAGAGTGCTGAAATGGGT | ACACACCCATCACAAGTCCA |
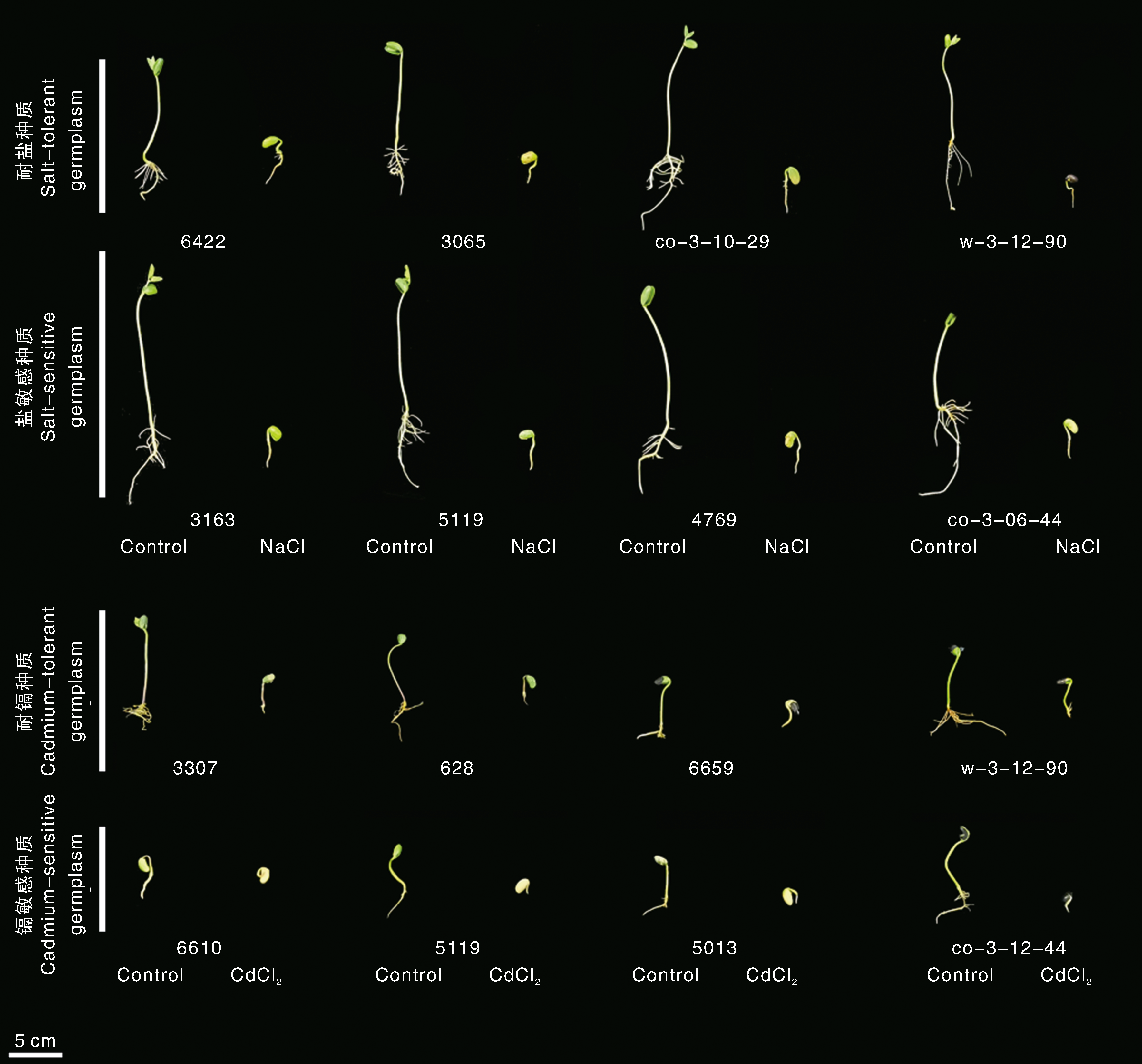
图1 不同大豆种质幼苗在盐或镉胁迫下的生理表现 盐、镉胁迫8 d后耐盐和耐镉品种,以及盐、镉敏感品种大豆幼苗的生理表现。NaCl,盐胁迫条件;CdCl2,镉胁迫条件;Control,正常生长条件。
Fig.1 Physiological performance of different soybean seedlings under salt or cadmium stress Physiological responses of soybean seedlings of salt/Cd-tolerant and salt/Cd-sensitive varieties under salt/Cd stress for 8 days. NaCl, Salt stress; Cd, Cadmium stress; Control, Normal growth.
| 处理 Treatment | PL/cm | RL/cm | RL/PL | SW/g | RW/g | GR | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 均值 Mean | CV/% | 均值 Mean | CV/% | 均值 Mean | CV/% | 均值 Mean | CV/% | 均值 Mean | CV/% | 均值 Mean | CV/% | |
| 正常条件 CK | 10.26± 0.31 a | 24.91 | 7.52± 0.16 a | 18.27 | 0.81± 0.02 b | 23.83 | 0.76± 0.02 a | 24.14 | 0.24± 0.01 a | 31.58 | 0.97± 0.01 a | 6.15 |
| 盐胁迫 Salt stress | 1.03± 0.02 b | 16.75 | 1.86± 0.05 b | 23.06 | 1.92± 0.05 a | 24.63 | 0.41± 0.01 b | 31.97 | 0.05± 0.00 b | 46.62 | 0.92± 0.01 b | 10.56 |
| 镉胁迫 Cd stress | 0.88± 0.03 b | 37.75 | 0.30± 0.04 c | 133.61 | 0.30± 0.04 c | 134.28 | 0.41± 0.01 b | 30.95 | 0.03± 0.01 b | 353.39 | 0.75± 0.02 c | 31.25 |
表2 50份大豆种质苗期在正常条件和盐/镉胁迫下的生理表现
Table 2 Physiological performance of 50 soybean germplasms during seedling stage under normal conditions and salt/cadmium stress
| 处理 Treatment | PL/cm | RL/cm | RL/PL | SW/g | RW/g | GR | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 均值 Mean | CV/% | 均值 Mean | CV/% | 均值 Mean | CV/% | 均值 Mean | CV/% | 均值 Mean | CV/% | 均值 Mean | CV/% | |
| 正常条件 CK | 10.26± 0.31 a | 24.91 | 7.52± 0.16 a | 18.27 | 0.81± 0.02 b | 23.83 | 0.76± 0.02 a | 24.14 | 0.24± 0.01 a | 31.58 | 0.97± 0.01 a | 6.15 |
| 盐胁迫 Salt stress | 1.03± 0.02 b | 16.75 | 1.86± 0.05 b | 23.06 | 1.92± 0.05 a | 24.63 | 0.41± 0.01 b | 31.97 | 0.05± 0.00 b | 46.62 | 0.92± 0.01 b | 10.56 |
| 镉胁迫 Cd stress | 0.88± 0.03 b | 37.75 | 0.30± 0.04 c | 133.61 | 0.30± 0.04 c | 134.28 | 0.41± 0.01 b | 30.95 | 0.03± 0.01 b | 353.39 | 0.75± 0.02 c | 31.25 |
| 性状 Trait | 主成分1 PC1 | 主成分2 PC2 | 主成分3 PC3 |
|---|---|---|---|
| PL | 0.86 | -0.12 | 0.08 |
| RL | 0.35 | 0.82 | -0.12 |
| GR | -0.49 | 0.84 | -0.09 |
| RL/PL | 0.42 | 0.67 | 0.24 |
| RW | 0.84 | -0.04 | 0.20 |
| SW | -0.37 | 0.04 | 0.90 |
| 特征值Eigenvalue | 2.09 | 1.81 | 0.93 |
| 贡献率/%Contribution rate/% | 34.85 | 30.12 | 15.52 |
| 累计贡献率/% | 34.85 | 64.97 | 80.49 |
| Cumulative contribution rate/% |
表3 50份大豆种质响应盐胁迫各变量对主成分的贡献度
Table 3 Contribution of variables to principal components under salt stress across 50 soybean germplasms
| 性状 Trait | 主成分1 PC1 | 主成分2 PC2 | 主成分3 PC3 |
|---|---|---|---|
| PL | 0.86 | -0.12 | 0.08 |
| RL | 0.35 | 0.82 | -0.12 |
| GR | -0.49 | 0.84 | -0.09 |
| RL/PL | 0.42 | 0.67 | 0.24 |
| RW | 0.84 | -0.04 | 0.20 |
| SW | -0.37 | 0.04 | 0.90 |
| 特征值Eigenvalue | 2.09 | 1.81 | 0.93 |
| 贡献率/%Contribution rate/% | 34.85 | 30.12 | 15.52 |
| 累计贡献率/% | 34.85 | 64.97 | 80.49 |
| Cumulative contribution rate/% |
| 性状 Trait | 主成分1 PC1 | 主成分2 PC2 | 主成分3 PC3 |
|---|---|---|---|
| PL | 0.76 | -0.19 | 0.52 |
| RL | 0.94 | 0.25 | -0.16 |
| RL/PL | 0.55 | -0.30 | -0.14 |
| GR | 0.65 | 0.53 | -0.51 |
| RW | 0.62 | -0.48 | 0.20 |
| SW | 0.05 | 0.70 | 0.66 |
| 特征值Eigenvalue | 2.56 | 1.20 | 1.05 |
| 贡献率/%Contribution rate/% | 42.68 | 20.01 | 17.51 |
| 累计贡献率/% | 42.68 | 62.69 | 80.20 |
| Cumulative contribution rate/% |
表4 50份大豆种质响应镉胁迫各变量对主成分的贡献度
Table 4 Contribution of variables to principal components under cadmium stress across 50 soybean germplasms
| 性状 Trait | 主成分1 PC1 | 主成分2 PC2 | 主成分3 PC3 |
|---|---|---|---|
| PL | 0.76 | -0.19 | 0.52 |
| RL | 0.94 | 0.25 | -0.16 |
| RL/PL | 0.55 | -0.30 | -0.14 |
| GR | 0.65 | 0.53 | -0.51 |
| RW | 0.62 | -0.48 | 0.20 |
| SW | 0.05 | 0.70 | 0.66 |
| 特征值Eigenvalue | 2.56 | 1.20 | 1.05 |
| 贡献率/%Contribution rate/% | 42.68 | 20.01 | 17.51 |
| 累计贡献率/% | 42.68 | 62.69 | 80.20 |
| Cumulative contribution rate/% |
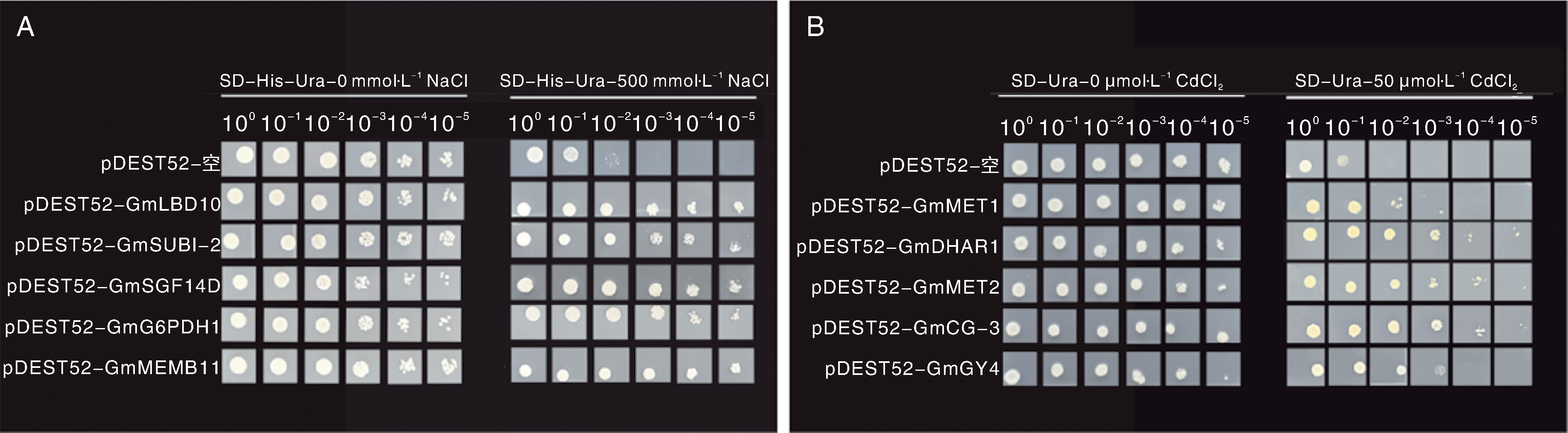
图2 转化大豆文库的酵母菌株在不同生长条件下的生长表现 A,大豆全长cDNA文库转化酵母G19菌株在含500 mmol·L-1 NaCl的SD-His-Ura培养基上的存活情况;B,大豆全长cDNA文库转化酵母Δycf1菌株在含50 μmol·L-1 CdCl2的SD-Ura培养基上的存活情况。照片拍摄于点板后第3天。
Fig.2 Growth performance of yeast strains transformed with soybean library under different growth conditions A, The survival performance of yeast strain G19 transformed with full-length soybean cDNA library on SD-His-Ura medium supplemented with 500 mmol·L-1 NaCl; B, The survival performance of yeast strain Δycf1 transformed with full-length soybean cDNA library on SD-Ura medium supplemented with 50 μmol·L-1 CdCl2. The photos were taken three days after plating.
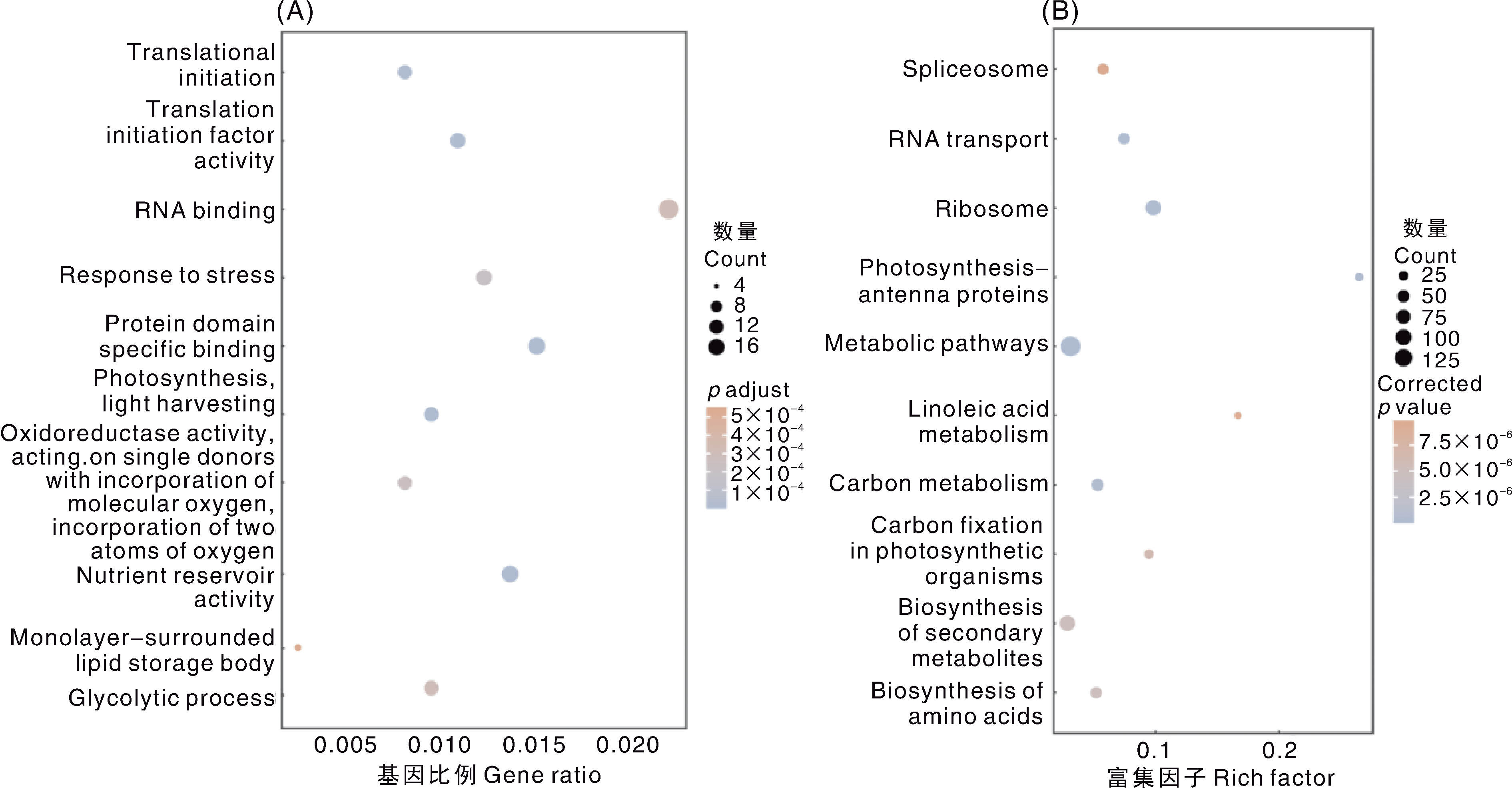
图3 大豆耐盐候选基因的GO、KEGG分析 A,GO功能聚类分析;B,KEGG通路富集分析。A图注释名称自上而下依次为:翻译起始;翻译起始因子活性;RNA结合;胁迫响应;蛋白质结构域特异性结合;光合作用,光捕获;氧化还原酶活性,作用于单一供体并结合分子氧,结合两个氧原子;营养储存活性;单层膜包被的脂质储存体;糖酵解过程。B图通路名称自上而下依次为:剪接体;RNA转运;核糖体;光合作用-天线蛋白;代谢途径;亚油酸代谢;碳代谢;光合生物中的碳固定;次生代谢物的生物合成;氨基酸的生物合成。
Fig.3 GO and KEGG analysis of candidate genes for salt tolerance in soybean A, GO function clustering analysis; B, KEGG pathway enrichment analysis. The annotation names of Fig. A from top to bottom are as follows: Translational initiation; Translation initiation factor activity; RNA binding; Response to stress; Protein domain specific binding; Photosynthesis, light harvesting; Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen; Nutrient reservoir activity; Monolayer-surrounded lipid storage body; Glycolytic process. The annotation names of Fig. B from top to bottom are as follows: Spliceosome; RNA transport; Ribosome; Photosynthesis-antenna proteins; Metabolic pathways; Linoleic acid metabolism; Carbon metabolism; Carbon fixation in photosynthetic organisms; Biosynthesis of secondary metabolites; Biosynthesis of amino acids.
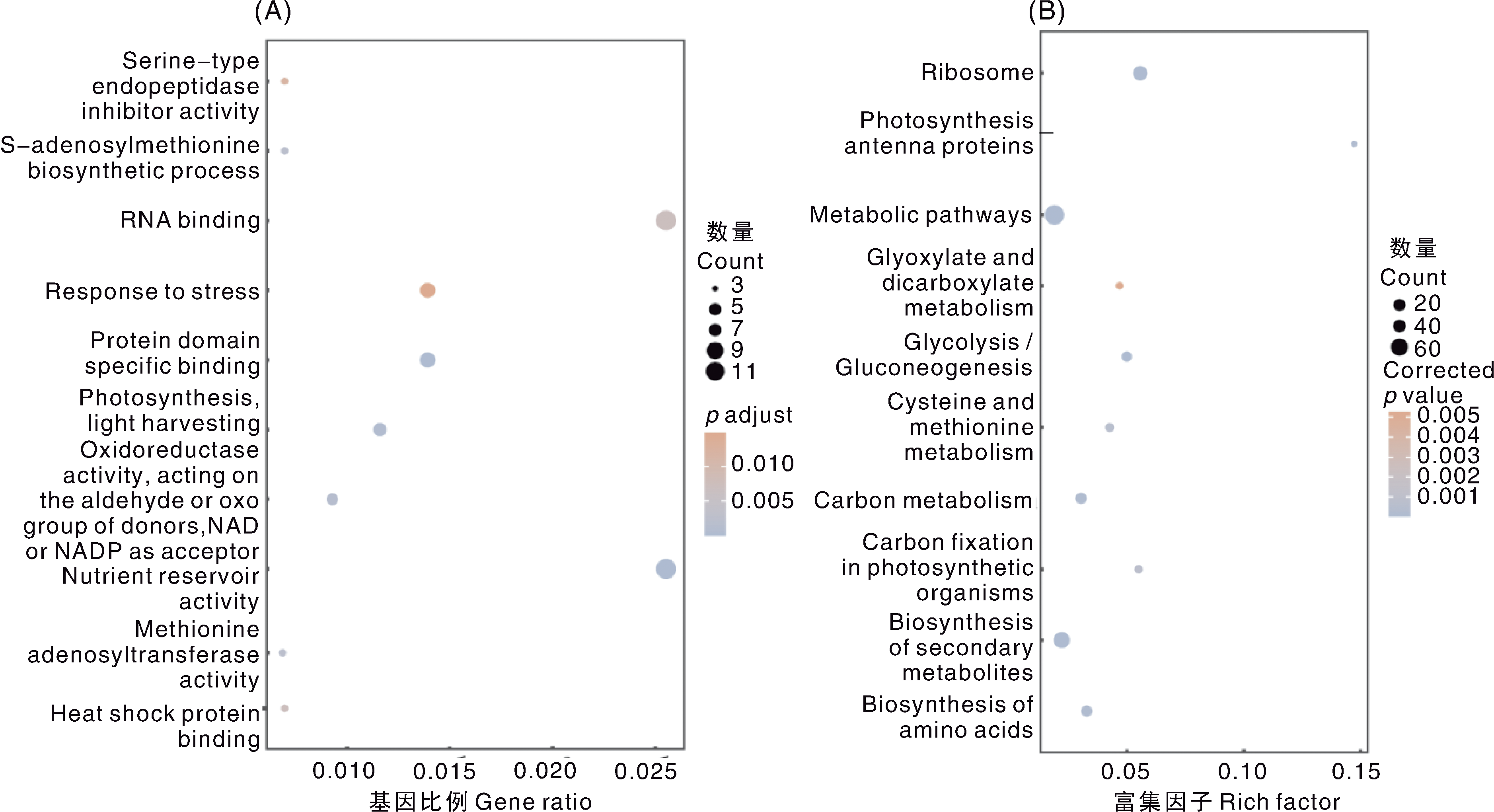
图4 大豆耐镉候选基因的GO、KEGG富集分析 A,GO功能的聚类分析;B,KEGG通路的富集分析。A图注释自上而下依次为:丝氨酸型内肽酶抑制活性;S-腺苷甲硫氨酸生物合成过程;RNA结合;胁迫响应;蛋白质结构域特异性结合;光合作用,光捕获;氧化还原酶活性(作用于供体的醛基或酮基,以NAD或NADP为受体);营养储存活性;甲硫氨酸腺苷转移酶活性;热休克蛋白结合。B图通路自上而下依次为:核糖体;光合作用-天线蛋白;代谢途径;乙醛酸和二羧酸代谢;糖酵解/糖异生;半胱氨酸和甲硫氨酸代谢;碳代谢;光合生物中的碳固定;次生代谢物生物合成;氨基酸生物合成。
Fig.4 GO and KEGG enrichment analysis of candidate genes for cadmium tolerance in soybean A, GO function clustering analysis; B, KEGG pathway enrichment analysis. The annotation names of Fig. A from top to bottom are as follows: Serine-type endopeptidase inhibitor activity; S-adenosylmethionine biosynthetic process; RNA binding; Response to stress; Protein domain specific binding; Photosynthesis, light harvesting; Oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor; Nutrient reservoir activity; Methionine adenosyltransferase activity; Heat shock protein binding. The annotation names of Fig. B from top to bottom are as follows: Ribosome; Photosynthesis-antenna proteins; Metabolic pathways; Glyoxylate and dicarboxylate metabolism; Glycolysis/Gluconeogenesis; Cysteine and methionine metabolism; Carbon metabolism; Carbon fixation in photosynthetic organisms; Biosynthesis of secondary metabolites; Biosynthesis of amino acids.
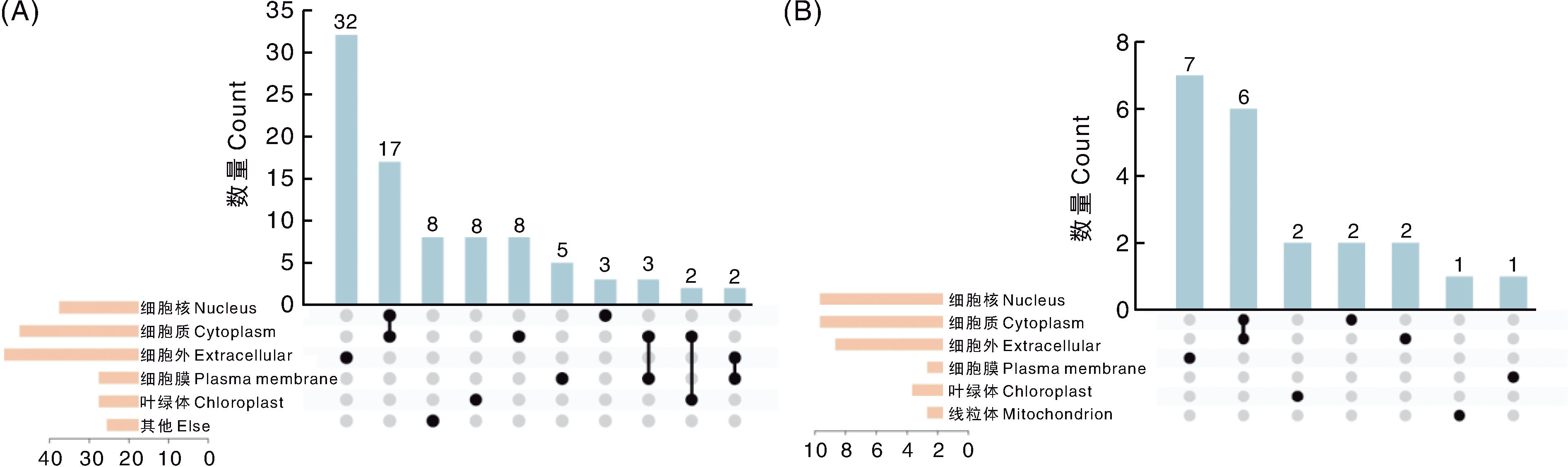
图5 大豆耐盐和耐镉候选基因编码蛋白的亚细胞定位 A,已知基因编码蛋白的亚细胞定位;B,未知基因编码蛋白的亚细胞定位。
Fig.5 Subcellular localization of proteins encoded by soybean salt and cadmium tolerance candidate genes A, Subcellular localization of proteins encoded by known genes; B, Subcellular localization of proteins encoded by unknown genes.
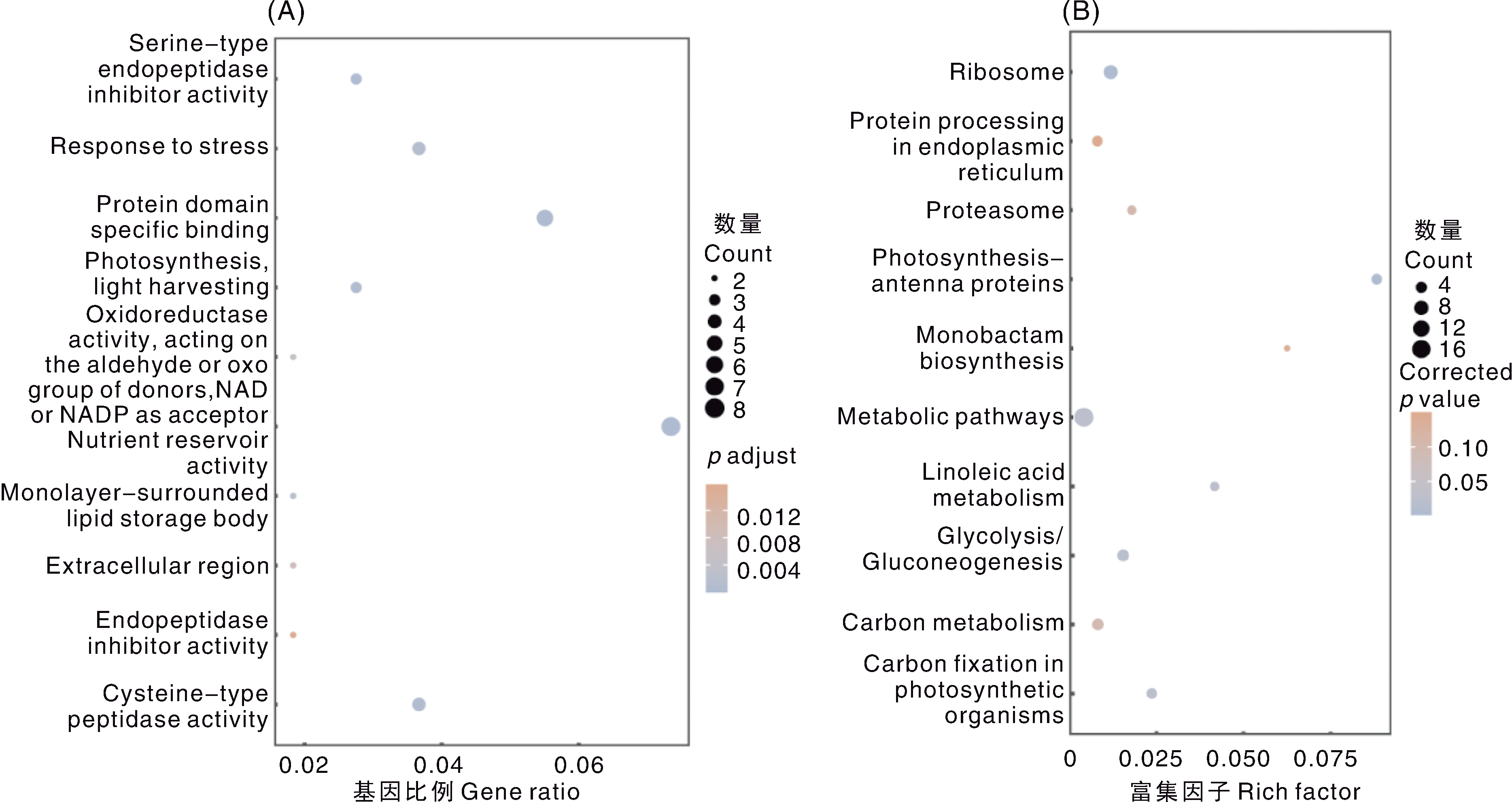
图6 大豆盐、镉共同耐性候选基因的GO、KEGG分析 A,GO功能的聚类分析;B,KEGG通路的富集分析。A图注释名称自上而下依次为:丝氨酸型内肽酶抑制活性;胁迫响应;蛋白质结构域特异性结合;光合作用,光捕获;氧化还原酶活性(作用于供体的醛基或酮基,以NAD或NADP 为受体);营养储存活性;单层膜包被的脂质储存体;细胞外区域;内肽酶抑制活性;半胱氨酸型肽酶活性。B图通路名称自上而下依次为:核糖体;内质网中的蛋白质加工;蛋白酶体;光合作用-天线蛋白;单环β-内酰胺生物合成;代谢途径;亚油酸代谢;糖酵解/糖异生;碳代谢;光合生物中的碳固定。Gene ratio表示注释到特定条目的差异基因数与差异基因总数的比值;Richfactor为某通路中富集到的差异基因与该通路所有基因的比值。下同。
Fig.6 GO and KEGG analysis of candidate genes for salt-cadmium co-tolerance in soybean A, GO function clustering analysis; B, KEGG pathway enrichment analysis. The annotation names of Fig. A from top to bottom are as follows: Serine-type endopeptidase inhibitor activity; Response to stress; Protein domain specific binding; Photosynthesis, light harvesting; Oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor; Nutrient reservoir activity; Monolayer-surrounded lipid storage body; Extracellular region; Endopeptidase inhibitor activity; Cysteine-type peptidase activity. The annotation names of Fig. B from top to bottom are as follows: Ribosome; Protein processing in endoplasmic reticulum; Proteasome; Photosynthesis-antenna proteins; Monobactam biosynthesis; Metabolic pathways; Linoleic acid metabolism; Glycolysis/Gluconeogenesis; Carbon metabolism; Carbon fixation in photosynthetic organisms. Gene ratio represents the ratio of differentially expressed genes annotated to a specific term to the total number of differentially expressed genes; Rich factor is the ratio of enriched differentially expressed genes in a pathway to all genes in that pathway. The same as below.
| 基因ID Gene ID | 基因名 Gene name | cDNA大小/bp cDNA size/bp | 预测功能 Function prediction |
|---|---|---|---|
| Glyma.09G185500 | GmDehydrin | 681 | Dehydrin-like protein |
| Glyma.03G163533 | GmSSP | 1 488 | Rmlc-like cupins superfamily protein |
| Glyma.02G208700 | GmGF14 | 789 | 14-3-3-like protein |
| Glyma.10G246300 | GmPAP85 | 1 866 | Cupin family protein |
| Glyma.15G072400 | GmHUP54 | 756 | Aluminium induced protein with YGL and LRDR motifs |
| Glyma.07G132000 | GmMET2 | 240 | Metallothionein 2A |
表5 大豆关键耐盐和镉基因信息
Table 5 Key salt and cadmium tolerant gene information of soybean
| 基因ID Gene ID | 基因名 Gene name | cDNA大小/bp cDNA size/bp | 预测功能 Function prediction |
|---|---|---|---|
| Glyma.09G185500 | GmDehydrin | 681 | Dehydrin-like protein |
| Glyma.03G163533 | GmSSP | 1 488 | Rmlc-like cupins superfamily protein |
| Glyma.02G208700 | GmGF14 | 789 | 14-3-3-like protein |
| Glyma.10G246300 | GmPAP85 | 1 866 | Cupin family protein |
| Glyma.15G072400 | GmHUP54 | 756 | Aluminium induced protein with YGL and LRDR motifs |
| Glyma.07G132000 | GmMET2 | 240 | Metallothionein 2A |
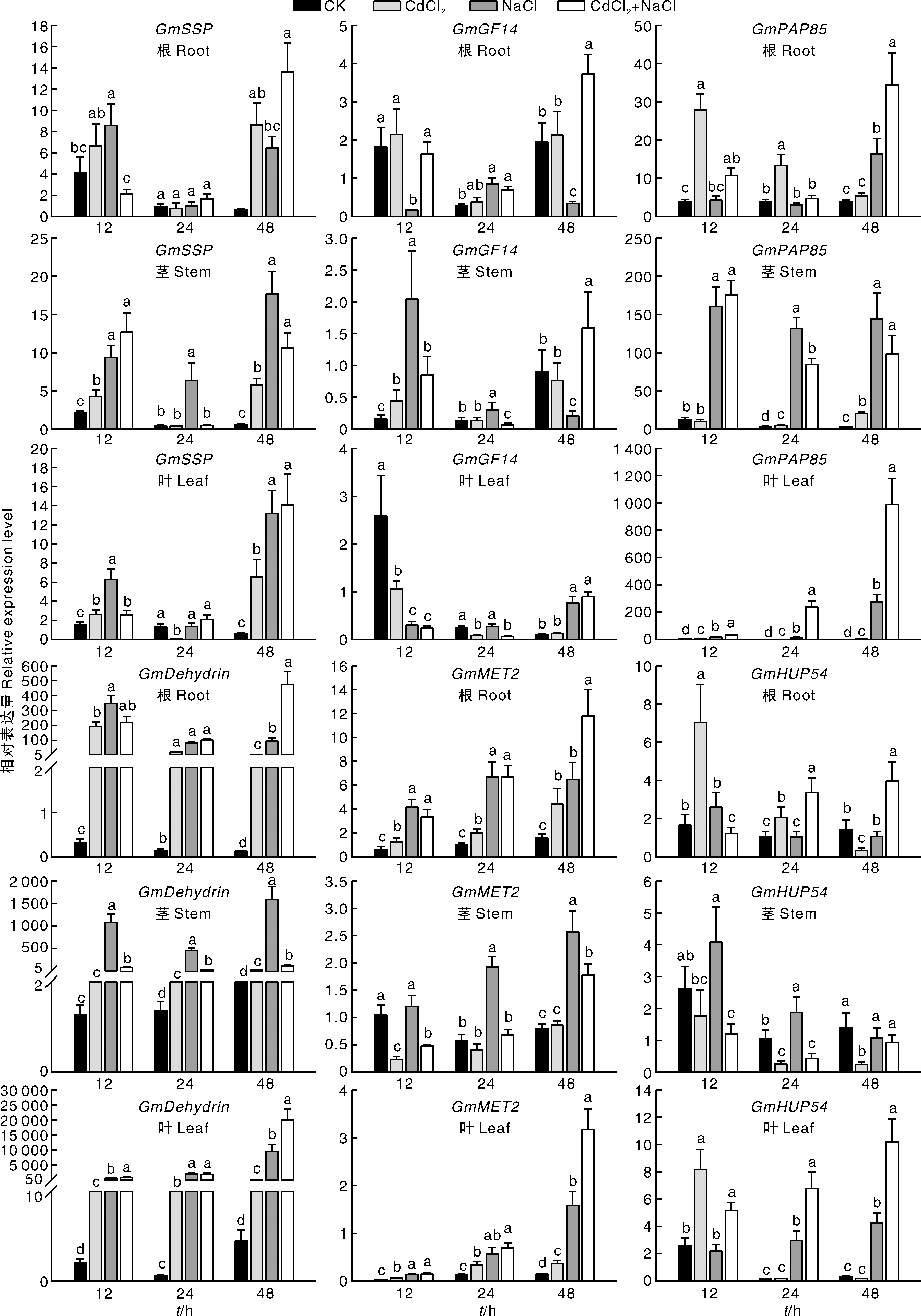
图7 盐、镉、盐+镉复合胁迫下大豆根、茎和叶中候选基因的相对表达水平 无相同小写字母表示同一个时间点在不同处理间差异显著(p<0.05)。
Fig.7 Relative expression level of candidate genes in soybean roots, stems, and leaves under salt, Cd and salt-Cd stress Bars marked with the same lowercase letters in the figure indicate significant differences (p<0.05) among treatments at the same time.
| [1] | HAMZA M, BASIT A W, SHEHZADI I, et al. Global impact of soybean production: a review[J]. Asian Journal of Biochemistry, Genetics and Molecular Biology, 2024, 16(2): 12-20. |
| [2] | 李奕聪, 杨钰莹, 李佳璇, 等. 2024年大豆产业发展趋势与政策建议[J]. 大豆科技, 2024(1): 1-5. |
| LI Y C, YANG Y Y, LI J X, et al. Development trends and policy suggestions of soybean industry in 2024[J]. Soybean Science & Technology, 2024(1): 1-5. | |
| [3] | 闫琰, 王秀东, 王济民, 等. “双循环” 背景下国家粮食安全战略研究[J]. 中国工程科学, 2023, 25(4): 14-25. |
| YAN Y, WANG X D, WANG J M, et al. National food security strategy against the backdrop of domestic and international dual circulation[J]. Strategic Study of CAE, 2023, 25(4): 14-25. | |
| [4] | SHANG C L, WANG L, TIAN C Y, et al. Heavy metal tolerance and potential for remediation of heavy metal-contaminated saline soils for the euhalophyte Suaeda salsa[J]. Plant Signaling & Behavior, 2020, 15(11): 1805902. |
| [5] | 唐希望, 周阳, 王龙雪, 等. 盐镉耦合胁迫对玉米种子萌发和幼苗生长的影响[J]. 东北农业科学, 2022, 47(5): 25-29. |
| TANG X W, ZHOU Y, WANG L X, et al. Effects of salt-cadmium coupling stress on seed germination and seedling growth of maize[J]. Journal of Northeast Agricultural Sciences, 2022, 47(5): 25-29. | |
| [6] | ZHANG W, LIAO X L, CUI Y M, et al. A cation diffusion facilitator, GmCDF1, negatively regulates salt tolerance in soybean[J]. PLoS Genetics, 2019, 15(1): e1007798. |
| [7] | SINGLETON P W, BEN BOHLOOL B. Effect of salinity on nodule formation by soybean[J]. Plant Physiology, 1984, 74(1): 72-76. |
| [8] | EL-SABAGH A, SOROUR S G R, UEDA A, et al. Evaluation of salinity stress effects on seed yield and quality of three soybean cultivars[J]. Azarian Journal of Agriculture, 2015, 2(5): 138-141. |
| [9] | DO T D, VUONG T D, DUNN D, et al. Mapping and confirmation of loci for salt tolerance in a novel soybean germplasm, Fiskeby Ⅲ[J]. Theoretical and Applied Genetics, 2018, 131(3): 513-524. |
| [10] | 黄运湘, 廖柏寒, 肖浪涛, 等. 镉处理对大豆幼苗生长及激素含量的影响[J]. 环境科学, 2006, 27(7): 1398-1401. |
| HUANG Y X, LIAO B H, XIAO L T, et al. Effects of Cd2+ on seedling growth and phytohormone contents of Glycine max[J]. Environmental Science, 2006, 27(7): 1398-1401. | |
| [11] | 陈朝明, 龚惠群, 王凯荣. Cd对桑叶品质、生理生化特性的影响及其机理研究[J]. 应用生态学报, 1996, 7(4): 417-423. |
| CHEN C M, GONG H Q, WANG K R. Effect of Cd on quality, physiological and biochemical characteristics of mulberry leaves and its mechanism[J]. Chinese Journal of Applied Ecology, 1996, 7(4): 417-423. | |
| [12] | LI J, GUO Y, YANG Y Q. The molecular mechanism of plasma membrane H+-ATPases in plant responses to abiotic stress[J]. Journal of Genetics and Genomics, 2022, 49(8): 715-725. |
| [13] | VAN ZELM E, ZHANG Y X, TESTERINK C. Salt tolerance mechanisms of plants[J]. Annual Review of Plant Biology, 2020, 71: 403-433. |
| [14] | FENG C, GAO H T, ZHOU Y G, et al. Unfolding molecular switches for salt stress resilience in soybean: recent advances and prospects for salt-tolerant smart plant production[J]. Frontiers in Plant Science, 2023, 14: 1162014. |
| [15] | AHMAD I, MIAN A, MAATHUIS F J M. Overexpression of the rice AKT1 potassium channel affects potassium nutrition and rice drought tolerance[J]. Journal of Experimental Botany, 2016, 67(9): 2689-2698. |
| [16] | DEINLEIN U, STEPHAN A B, HORIE T, et al. Plant salt-tolerance mechanisms[J]. Trends in Plant Science, 2014, 19(6): 371-379. |
| [17] | CHUNG Y S, KIM K S, HAMAYUN M, et al. Silicon confers soybean resistance to salinity stress through regulation of reactive oxygen and reactive nitrogen species[J]. Frontiers in Plant Science, 2020, 10: 1725. |
| [18] | 孙敏. 水稻植株中镉区室化关键螯合物的鉴定与分析[D]. 北京: 中国农业科学院, 2010. |
| SUN M. Characterization and analysis of chelates involved in compartmentation of cadmium in rice plants[D]. Beijing: Chinese Academy of Agricultural Sciences, 2010. | |
| [19] | 张旭红, 高艳玲, 林爱军, 等. 植物根系细胞壁在提高植物抵抗金属离子毒性中的作用[J]. 生态毒理学报, 2008, 3(1): 9-14. |
| ZHANG X H, GAO Y L, LIN A J, et al. A review on the effects of cell wall on the resistance of plants to metal stress[J]. Asian Journal of Ecotoxicology, 2008, 3(1): 9-14. | |
| [20] | HIRATA K, TSUJI N, MIYAMOTO K. Biosynthetic regulation of phytochelatins, heavy metal-binding peptides[J]. Journal of Bioscience and Bioengineering, 2005, 100(6): 593-599. |
| [21] | LUO P, WU J J, LI T T, et al. An overview of the mechanisms through which plants regulate ROS homeostasis under cadmium stress[J]. Antioxidants, 2024, 13(10): 1174. |
| [22] | FALLER P, KIENZLER K, KRIEGER-LISZKAY A. Mechanism of Cd2+ toxicity: Cd2+ inhibits photoactivation of Photosystem Ⅱ by competitive binding to the essential Ca2+ site[J]. Biochimica et Biophysica Acta(BBA)-Bioenergetics 2005, 1706(1/2): 158-164. |
| [23] | EL-SHINTINAWY F. Glutathione counteracts the inhibitory effect induced by cadmium on photosynthetic process in soybean[J]. Photosynthetica, 1999, 36(1): 171-179. |
| [24] | NING H X, ZHANG C H, YAO Y, et al. Overexpression of a soybean O-acetylserine (thiol) lyase-encoding gene GmOASTL4 in tobacco increases cysteine levels and enhances tolerance to cadmium stress[J]. Biotechnology Letters, 2010, 32(4): 557-564. |
| [25] | AZEVEDO R A, GRATÃO P L, MONTEIRO C C, et al. What is new in the research on cadmium-induced stress in plants?[J]. Food and Energy Security, 2012, 1(2): 133-140. |
| [26] | PESSARAKLI M. Handbook of plant and crop physiology[M]. 4th ed. Boca Raton: CRC Press, 2021. |
| [27] | UDDIN K M, JURAIMI A S, ISMAIL M R, et al. Relative salinity tolerance of warm season turfgrass species[J]. Journal of Environmental Biology, 2011, 32(3): 309-312. |
| [28] | CHEN Y, LI L L, ZONG J Q, et al. Heterologous expression of the halophyte Zoysia matrella H+-pyrophosphatase gene improved salt tolerance in Arabidopsis thaliana[J]. Plant Physiology and Biochemistry, 2015, 91: 49-55. |
| [29] | DALCORSO G, FARINATI S, FURINI A. Regulatory networks of cadmium stress in plants[J]. Plant Signaling & Behavior, 2010, 5(6): 663-667. |
| [30] | 韩毅强, 高亚梅, 杜艳丽, 等. 大豆耐盐碱种质资源鉴定[J]. 中国油料作物学报, 2021, 43(6): 1016-1024. |
| HAN Y Q, GAO Y M, DU Y L, et al. Identification of saline-alkali tolerant germplasm resources of soybean during the whole growth stage[J]. Chinese Journal of Oil Crop Sciences, 2021, 43(6): 1016-1024. | |
| [31] | 林峰, 赵慧艳, 史飞飞, 等. 大豆种质资源苗期耐盐鉴定及遗传多样性分析[J]. 植物遗传资源学报, 2024, 25(6): 945-956. |
| LIN F, ZHAO H Y, SHI F F, et al. Identification of salt-tolerant germplasm resources in soybean seedlings and genetic diversity analysis[J]. Journal of Plant Genetic Resources, 2024, 25(6): 945-956. | |
| [32] | ARAO T, ISHIKAWA S. Genotypic differences in cadmium concentration and distribution of soybean and rice[J]. Japan Agricultural Research Quarterly, 2006, 40(1): 21-30. |
| [33] | 周秀文, 张晓蕊, 孙贺祥, 等. 大豆种质萌发期和苗期耐盐性评价[J]. 沈阳农业大学学报, 2022, 53(3): 257-264. |
| ZHOU X W, ZHANG X R, SUN H X, et al. Evaluation of salt tolerance of soybean germplasms at germination and seedling stages[J]. Journal of Shenyang Agricultural University, 2022, 53(3): 257-264. | |
| [34] | 严勇亮, 张金波, 路子峰, 等. 大豆种质资源耐盐性鉴定与评价[J]. 新疆农业科学, 2021, 58(1): 65-71. |
| YAN Y L, ZHANG J B, LU Z F, et al. Salt tolerance evaluation of soybean germplasm[J]. Xinjiang Agricultural Sciences, 2021, 58(1): 65-71. | |
| [35] | SHAMSI I H, ZHANG G P, HU H L, et al. Assessment of the hazardous effects of Cd on physiological and biochemical characteristics of soybean genotypes[J]. International Journal of Agriculture and Biology, 2014, 16(1): 41-48. |
| [36] | ZHI Y, SUN T, ZHOU Q X, et al. Screening of safe soybean cultivars for cadmium contaminated fields[J]. Scientific Reports, 2020, 10: 12965. |
| [37] | 石广成. 大豆耐盐种质筛选及GmSCAMPs家族的耐盐功能分析[D]. 太谷: 山西农业大学, 2022. |
| SHI G C. Screening of soybean salt-tolerant germplasm and study on salt-tolerant function of GmSCAMPs family[D]. Taigu: Shanxi Agricultural University, 2022. | |
| [38] | 袁宇婷, 张晓燕, 吴谷丰, 等. 基于主成分和隶属函数分析的大豆种质资源耐盐性综合评价[J]. 大豆科学, 2025, 44(1): 22-32. |
| YUAN Y T, ZHANG X Y, WU G F, et al. Comprehensive evaluation of salt tolerance of soybean germplasm resources based on principal component and membership function analysis[J]. Soybean Science, 2025, 44(1): 22-32. | |
| [39] | 张新草, 薛项潇, 姜深, 等. 大豆种质发芽期耐盐碱性鉴定及指标筛选[J]. 西北农业学报, 2020, 29(3): 374-381. |
| ZHANG X C, XUE X X, JIANG S, et al. Identification of mixed saline-alkali tolerance and screening of indicators in soybean at germination stage[J]. Acta Agriculturae Boreali-occidentalis Sinica, 2020, 29(3): 374-381. | |
| [40] | 刘佳丽. 重金属镉对大豆种子萌发与幼苗生长的影响[J]. 农业技术与装备, 2023(12): 13-15. |
| LIU J L. Effects of heavy metal cadmium on seed germination and seedling growth of soybean[J]. Agricultural Technology & Equipment, 2023(12): 13-15. | |
| [41] | 郭远, 王文成, 徐颖莹, 等. 植物耐盐评价方法综述[J]. 江苏农业科学, 2017, 45(23): 18-23. |
| GUO Y, WANG W C, XU Y Y, et al. Review on evaluation methods of plant salt tolerance[J]. Jiangsu Agricultural Sciences, 2017, 45(23): 18-23. | |
| [42] | TAO J Y, LU L L, TAO J Y, et al. Advances in genes-encoding transporters for cadmium uptake, translocation, and accumulation in plants[J]. Toxics, 2022, 10(8): 411. |
| [43] | SHIRAKU M L, MAGWANGA R O, ZHANG Y Y, et al. Late embryogenesis abundant gene LEA3(Gh_A08G0694) enhances drought and salt stress tolerance in cotton[J]. International Journal of Biological Macromolecules, 2022, 207: 700-714. |
| [44] | LI X, FENG H, LIU S, et al. Dehydrin CaDHN2 enhances drought tolerance by affecting ascorbic acid synthesis under drought in peppers[J]. Plants, 2023, 12(22): 3895. |
| [45] | PAL P, MASAND M, SHARMA S, et al. Genome-wide transcriptional profiling and physiological investigation elucidating the molecular mechanism of multiple abiotic stress response in Stevia rebaudiana Bertoni[J]. Scientific Reports, 2023, 13: 19853. |
| [46] | SHIRAKU M L, MAGWANGA R O, CAI X Y, et al. Knockdown of 60S ribosomal protein L14-2 reveals their potential regulatory roles to enhance drought and salt tolerance in cotton[J]. Journal of Cotton Research, 2021, 4(1): 27. |
| [47] | GONG J W, LIU Q, CAI L L, et al. Multimechanism collaborative superior antioxidant CDzymes to alleviate salt stress-induced oxidative damage in plant growth[J]. ACS Sustainable Chemistry & Engineering, 2023, 11(10): 4237-4247. |
| [48] | CHUNG E S, KIM K Y, SO H A, et al. Enhanced tolerance against osmotic stresses of Escherichia coli cells expressing soybean KS-type dehydrin[J]. Genes & Genomics, 2008, 30(4): 319-327. |
| [49] | ŠVECOVÁ M, BOSZORÁDOVÁ E, MATUŠÍKOVÁ I, et al. Arabidopsis AtLTI30 and AtHIRD11 dehydrin genes and their contribution to cadmium tolerance in transgenic tobacco plants[J]. Acta Physiologiae Plantarum, 2022, 45(2): 21. |
| [50] | AHAMMED G J, LI C X, LI X, et al. Overexpression of tomato RING E3 ubiquitin ligase gene SlRING1 confers cadmium tolerance by attenuating cadmium accumulation and oxidative stress[J]. Physiologia Plantarum, 2021, 173(1): 449-459. |
| [51] | LIU J, QI W C, LU H Y, et al. Characterization of interactions between the soybean salt-stress responsive membrane-intrinsic proteins GmPIP1 and GmPIP2[J]. Agronomy, 2021, 11(7): 1312. |
| [52] | ZHOU L, WANG C, LIU R F, et al. Constitutive overexpression of soybean plasma membrane intrinsic protein GmPIP1;6 confers salt tolerance[J]. BMC Plant Biology, 2014, 14: 181. |
| [53] | HU W, YUAN Q Q, WANG Y, et al. Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic tobacco[J]. Plant & Cell Physiology, 2012, 53(12): 2127-2141. |
| [54] | PAGANI M A, TOMAS M, CARRILLO J, et al. The response of the different soybean metallothionein isoforms to cadmium intoxication[J]. Journal of Inorganic Biochemistry, 2012, 117: 306-315. |
| [55] | MANOSALVA P M, BRUCE M, LEACH J E. Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance[J]. Plant Journal, 2011, 68(5): 777-787. |
| [56] | QIU W M, SONG X X, HAN X J, et al. Overexpression of Sedum alfredii cinnamyl alcohol dehydrogenase increases the tolerance and accumulation of cadmium in Arabidopsis[J]. Environmental and Experimental Botany, 2018, 155: 566-577. |
| [57] | ZHANG J, YANG N, LI Y Y, et al. Overexpression of PeMIPS1 confers tolerance to salt and copper stresses by scavenging reactive oxygen species in transgenic poplar[J]. Tree Physiology, 2018, 38(10): 1566-1577. |
| [1] | 胡莹洁, 杜晨琪, 王鎏帆, 寿建昕, 王超, 徐梅, 严旭. 囊泡运输调控植物盐胁迫响应的研究进展[J]. 浙江农业学报, 2025, 37(9): 2003-2011. |
| [2] | 关秀生, 刘铁山, 王娟, 张茂林, 刘春晓, 董瑞, 关海英, 刘强, 徐扬, 何春梅. 玉米NF-YA家族基因的生物信息学分析与克隆[J]. 浙江农业学报, 2025, 37(8): 1605-1614. |
| [3] | 何国欣, 李素娟, 王剑, 陶晓园, 叶子弘, 陈光, 徐盛春. 大豆种质苗期低氮耐性筛选和鉴定[J]. 浙江农业学报, 2025, 37(5): 965-976. |
| [4] | 许竹溦, 雷俊, 邵晓伟, 陈润兴, 姜欢, 汪寿根, 余文慧. 基于层次分析法与模糊综合评价法的衢州鲜食大豆低聚糖种质资源评价研究[J]. 浙江农业学报, 2025, 37(4): 754-766. |
| [5] | 汤奥冉, 金秀, 王坦, 饶元, 李佳佳, 张武. 基于弯曲大豆植株主茎骨架重构的生理株高测量方法[J]. 浙江农业学报, 2025, 37(2): 466-479. |
| [6] | 廖小龙, 王兴胜, 陈勇, 李斌, 洪思丹, 梅利那, 国颖. 杨属植物HKT基因家族成员鉴定与盐胁迫下的表达模式分析[J]. 浙江农业学报, 2025, 37(10): 2104-2115. |
| [7] | 欧晋稳, 张古文, 冯志娟, 王斌, 卜远鹏, 徐钰, 茹磊, 刘娜, 龚亚明. 大豆海藻糖-6-磷酸磷酸酶基因GmTPP的鉴定及其在生长发育和非生物胁迫响应中的表达分析[J]. 浙江农业学报, 2024, 36(9): 2031-2041. |
| [8] | 彭佳诚, 吴越, 徐洁皓, 夏美文, 齐天鹏, 徐海圣. 日本沼虾桩蛋白基因的克隆与镉胁迫对其表达的影响[J]. 浙江农业学报, 2024, 36(2): 247-253. |
| [9] | 高憬, 陆玲鸿, 古咸彬, 范飞, 宋根华, 张慧琴. 猕猴桃AcWRKY94基因的克隆及其在盐胁迫下的功能分析[J]. 浙江农业学报, 2024, 36(11): 2501-2509. |
| [10] | 唐跃辉, 陈淑颖, 何文琼, 王涵瑾, 包欣欣, 贾赛男, 王瑶瑶, 陈宇阳, 杨同文. 麻风树JcERF22基因的克隆与功能分析[J]. 浙江农业学报, 2024, 36(10): 2219-2228. |
| [11] | 孙秀娟, 徐伟慧, 王志刚. 大豆根瘤内生细菌的分离鉴定及其对大豆植株的促生效应[J]. 浙江农业学报, 2023, 35(7): 1532-1541. |
| [12] | 卜远鹏, 刘娜, 张古文, 冯志娟, 王斌, 龚亚明, 许林英. 菜用大豆种质资源的农艺性状多样性评价及核心种质与食味品质评价体系的构建[J]. 浙江农业学报, 2023, 35(6): 1307-1314. |
| [13] | 杨松花, 石贵阳, 王晶琴, 陈竹. 低磷胁迫下大豆根系分泌物对土壤中难溶性磷的影响[J]. 浙江农业学报, 2023, 35(6): 1396-1406. |
| [14] | 檀舒霞, 赵桃弟, 杨豪, 宁可君, 刘丽, 何庆元, 黄守程, 舒英杰. 遮阴对10个菜用大豆品种农艺性状、产量和硝态氮代谢的影响[J]. 浙江农业学报, 2023, 35(4): 729-735. |
| [15] | 张梦, 佘宝, 杨玉莹, 黄林生, 朱梦琦. 基于无人机RGB影像的大豆种植区提取方法研究[J]. 浙江农业学报, 2023, 35(4): 952-961. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

