浙江农业学报 ›› 2026, Vol. 38 ›› Issue (1): 17-23.DOI: 10.3969/j.issn.1004-1524.20250089
利用CRISPR/Cas9技术创制糯稻新种质
潘月云( ), 黄雨青, 丁正权, 施扬, 黄海祥, 李白(
), 黄雨青, 丁正权, 施扬, 黄海祥, 李白( )
)
- 嘉兴市农业科学研究院, 浙江 嘉兴 314016
-
收稿日期:2025-02-07出版日期:2026-01-25发布日期:2026-02-11 -
作者简介:李白,E-mail:libaia@yeah.net
潘月云,从事水稻育种工作。E-mail:panyueyun3160@163.com -
通讯作者:李白 -
基金资助:嘉兴市科技计划项目(2023AZ11007);嘉兴市科技计划项目(2024AZ10003)
Generation of new glutinous rice germplasms via CRISPR/Cas9 technology
PAN Yueyun( ), HUANG Yuqing, DING Zhengquan, SHI Yang, HUANG Haixiang, LI Bai(
), HUANG Yuqing, DING Zhengquan, SHI Yang, HUANG Haixiang, LI Bai( )
)
- Jiaxing Academy of Agricultural Sciences, Jiaxing 314016, Zhejiang, China
-
Received:2025-02-07Online:2026-01-25Published:2026-02-11 -
Contact:LI Bai
摘要:
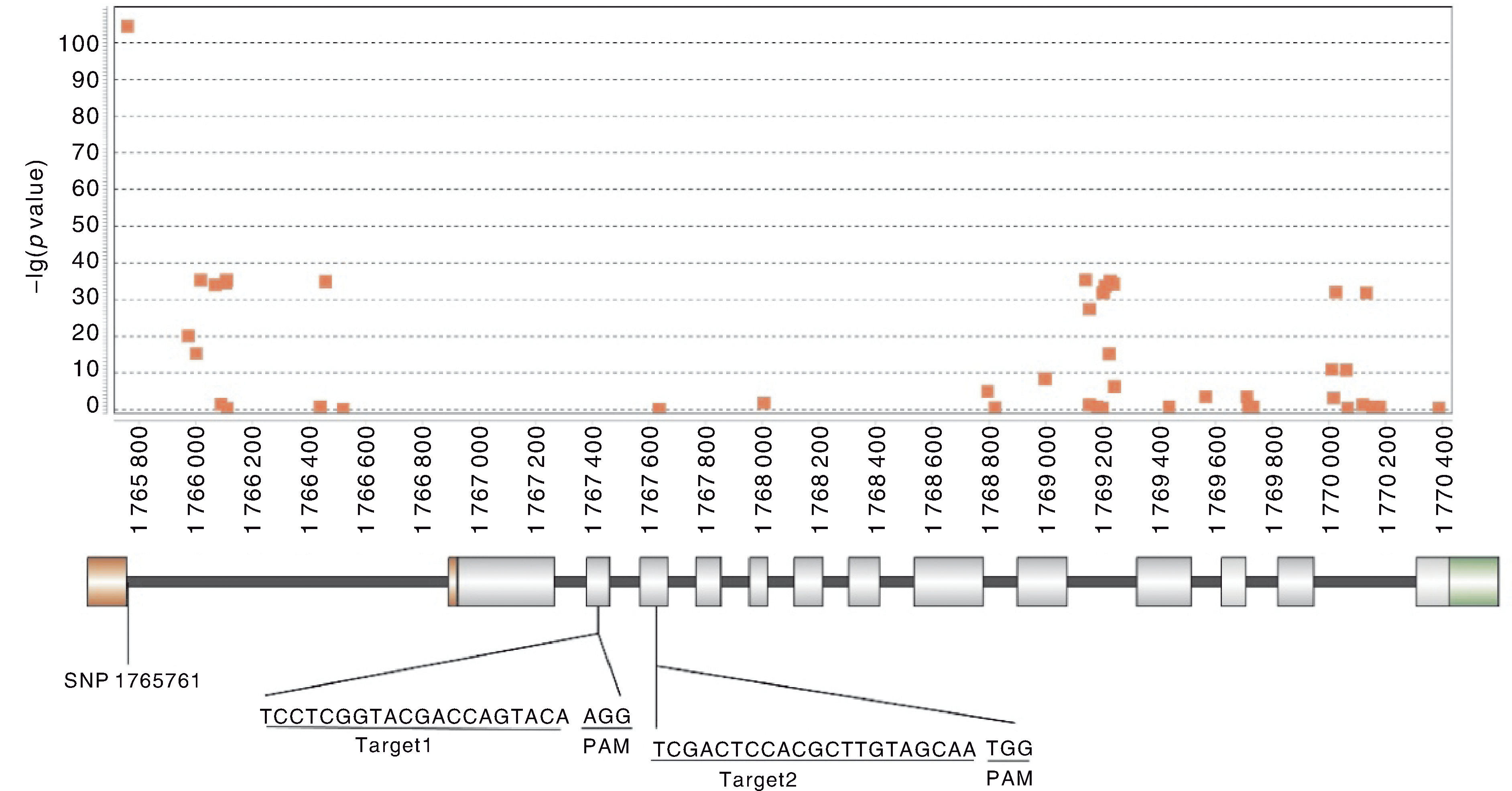
稻米直链淀粉含量直接影响其蒸煮食味品质,该性状主要由Waxy(Wx)基因控制。本研究基于Wx基因上SNP的表型关联分析,选择其第3、第4外显子设计编辑靶点。利用CRISPR/Cas9技术,以水稻品种嘉禾香1号为受体,对Wx基因进行编辑。经PCR与Sanger测序验证,获得4个不同类型的双位点纯合突变体。4个双位点纯合T2代突变体株系的稻米均呈蜡质状,其直链淀粉含量由野生型的16.9%降至0.83%~1.23%,达到糯稻水平。结果表明,利用CRISPR/Cas9技术可快速创制优质香糯型水稻种质资源。
中图分类号:
引用本文
潘月云, 黄雨青, 丁正权, 施扬, 黄海祥, 李白. 利用CRISPR/Cas9技术创制糯稻新种质[J]. 浙江农业学报, 2026, 38(1): 17-23.
PAN Yueyun, HUANG Yuqing, DING Zhengquan, SHI Yang, HUANG Haixiang, LI Bai. Generation of new glutinous rice germplasms via CRISPR/Cas9 technology[J]. Acta Agriculturae Zhejiangensis, 2026, 38(1): 17-23.
| 引物名称 Primer name | 引物序列(5'→3') Primer sequence(5'→3') |
|---|---|
| Wx_sg1F | ttttggtctcctgcaTCCTCGGTACGACCAGTACAGTTTC- AGAGCTATGCTGGAAAC |
| Wx_sg2R | attcggtctccaaacTTGCTACAAGCGTGGAGTCGTGC- ACCAGCCGGGAATCGAA |
| Cas9_755F | GCCTACCACGAGAAGTACCC |
| Cas9_755R | GGCTTGATGAACTTGTAGAACTC |
| Wx_seqF | GCTGTAAGCACACACAAACTTC |
| Wx_seqR | AGAAGTAATCACTGACCTGGC |
表1 引物序列
Table 1 Primer sequences
| 引物名称 Primer name | 引物序列(5'→3') Primer sequence(5'→3') |
|---|---|
| Wx_sg1F | ttttggtctcctgcaTCCTCGGTACGACCAGTACAGTTTC- AGAGCTATGCTGGAAAC |
| Wx_sg2R | attcggtctccaaacTTGCTACAAGCGTGGAGTCGTGC- ACCAGCCGGGAATCGAA |
| Cas9_755F | GCCTACCACGAGAAGTACCC |
| Cas9_755R | GGCTTGATGAACTTGTAGAACTC |
| Wx_seqF | GCTGTAAGCACACACAAACTTC |
| Wx_seqR | AGAAGTAATCACTGACCTGGC |
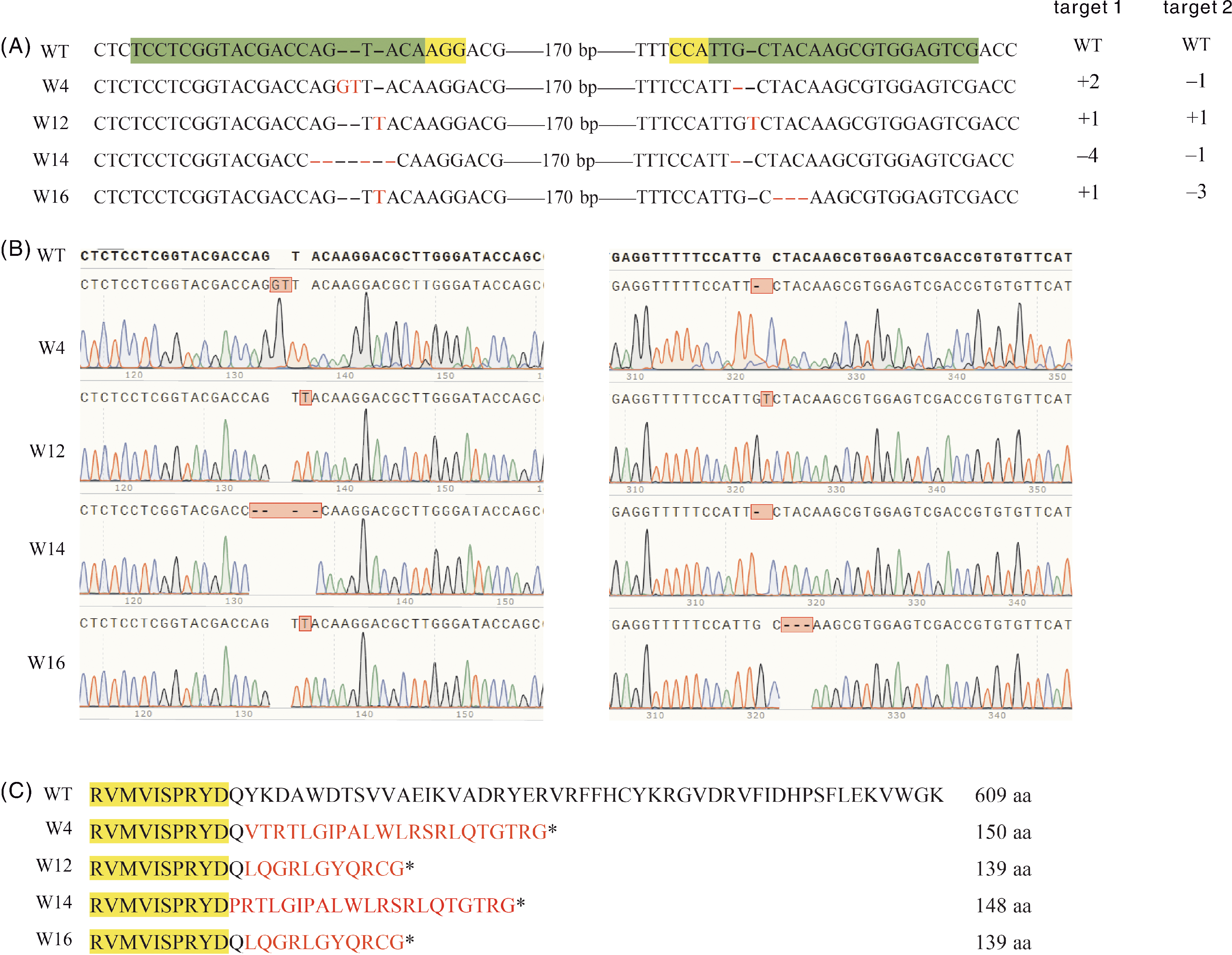
图3 4种纯合突变系的突变类型 A,纯合突变体的基因型,靶序列以绿色背景标示,PAM序列以黄色背景标示,变异碱基以红色标示。B,测序峰图。C,氨基酸序列的变异,变异氨基酸以红色标示。WT,野生型;W4、W12、W14和W16为双位点纯合突变体株系。
Fig.3 Mutation types of the four homozygous mutant lines A, The mutant genotype of the homozygous lines, the targeted sequence is highlighted in green shadow and the PAM sequence in yellow shadow, the changed nucleotides are highlighted in red. B, Sequencing peak map of the target region. C, Amino acid sequence of the mutant lines, the changed amino acids are highlighted in red. WT, Wild type; W4, W12, W14 and W16 are biallelic homozygous mutant lines.

图4 野生型与纯合突变株系籽粒表型 WT,野生型;W4、W12、W14和W16为双位点纯合突变体株系。
Fig.4 Grain phenotypes of wild-type and homozygous mutant lines WT, Wild type; W4, W12, W14 and W16 are biallelic homozygous mutant lines.
| 株系 Line | 株高/cm Plant height/cm | 粒长/mm Grain length/mm | 粒宽/mm Grain width/mm | 结实率/% Seed setting rate/% | 千粒重/g 1 000-grain weight/g | 单株有效穗数 Panicle number per plant | 每穗粒数 Grain number per panicle |
|---|---|---|---|---|---|---|---|
| WT | 102.15±3.66 | 10.54±0.08 | 3.51±0.05 | 85.33±3.13 | 30.61±0.84 | 10.70±1.06 | 145.50±8.64 |
| W4 | 101.85±3.91 | 10.13±0.07 | 3.45±0.04 | 84.93±2.91 | 30.13±0.98 | 10.60±0.97 | 143.60±9.31 |
| W12 | 102.78±3.40 | 10.64±0.08 | 3.50±0.06 | 85.88±3.04 | 30.65±0.77 | 10.40±0.84 | 143.45±9.02 |
| W14 | 99.58±2.87 | 10.38±0.09 | 3.49±0.06 | 86.45±3.78 | 29.64±1.02 | 10.30±1.34 | 139.73±12.83 |
| W16 | 99.85±3.54 | 9.98±0.07 | 3.42±0.05 | 83.86±2.06 | 29.39±0.87 | 10.80±1.14 | 147.38±9.40 |
表2 野生型与突变体的农艺性状
Table 2 Agronomic traits of wild-type and homozygous mutant lines
| 株系 Line | 株高/cm Plant height/cm | 粒长/mm Grain length/mm | 粒宽/mm Grain width/mm | 结实率/% Seed setting rate/% | 千粒重/g 1 000-grain weight/g | 单株有效穗数 Panicle number per plant | 每穗粒数 Grain number per panicle |
|---|---|---|---|---|---|---|---|
| WT | 102.15±3.66 | 10.54±0.08 | 3.51±0.05 | 85.33±3.13 | 30.61±0.84 | 10.70±1.06 | 145.50±8.64 |
| W4 | 101.85±3.91 | 10.13±0.07 | 3.45±0.04 | 84.93±2.91 | 30.13±0.98 | 10.60±0.97 | 143.60±9.31 |
| W12 | 102.78±3.40 | 10.64±0.08 | 3.50±0.06 | 85.88±3.04 | 30.65±0.77 | 10.40±0.84 | 143.45±9.02 |
| W14 | 99.58±2.87 | 10.38±0.09 | 3.49±0.06 | 86.45±3.78 | 29.64±1.02 | 10.30±1.34 | 139.73±12.83 |
| W16 | 99.85±3.54 | 9.98±0.07 | 3.42±0.05 | 83.86±2.06 | 29.39±0.87 | 10.80±1.14 | 147.38±9.40 |
| [1] | ZHOU Z K, ROBARDS K, HELLIWELL S, et al. Composition and functional properties of rice[J]. International Journal of Food Science and Technology, 2002, 37(8): 849-868. |
| [2] | JULIANO B O. Structure, chemistry, and function of the rice grain and its fractions[J]. Cereal Foods World, 1992, 37: 772-779. |
| [3] | 齐琦. 黑糯米保健酒的工艺及品质研究[D]. 贵阳: 贵州大学, 2018. |
| QI Q. Study on the technology and quality of black glutinous rice health wine[D]. Guiyang: Guizhou University, 2018. | |
| [4] | 龙夫. 中国少数民族的美容文化[J]. 中国民族博览, 2000(2): 46-47. |
| LONG F. The beauty culture of ethnic minorities in China[J]. China National Exhibition, 2000(2): 46-47. | |
| [5] | WANG Z Y, WU Z L, XING Y Y, et al. Nucleotide sequence of rice waxy gene[J]. Nucleic Acids Research, 1990, 18(19): 5898. |
| [6] | WANG Z Y, ZHENG F Q, SHEN G Z, et al. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene[J]. Plant Journal, 1995, 7(4): 613-622. |
| [7] | ISSHIKI M, MORINO K, NAKAJIMA M, et al. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5' splice site of the first intron[J]. Plant Journal, 1998, 15(1): 133-138. |
| [8] | WANCHANA S, TOOJINDA T, TRAGOONRUNG S, et al. Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.)[J]. Plant Science, 2003, 165(6): 1193-1199. |
| [9] | YANG J, WANG J, FAN F J, et al. Development of AS-PCR marker based on a key mutation confirmed by resequencing of Wx-mp in Milky Princess and its application in japonica soft rice (Oryza sativa L.) breeding[J]. Plant Breeding, 2013, 132(6): 595-603. |
| [10] | SATO H, SUZUKI Y, SAKAI M, et al. Molecular characterization of wx-mq, a novel mutant gene for low-amylose content in endosperm of rice (Oryza sativa L.)[J]. Breeding Science, 2002, 52(2): 131-135. |
| [11] | YANG Y, ZHOU L H, FENG L H, et al. Deciphering the role of waxy gene mutations in enhancing rice grain quality[J]. Foods, 2024, 13(11): 1624. |
| [12] | ZHANG C Q, ZHU J H, CHEN S J, et al. Wx lv, the ancestral allele of rice waxy gene[J]. Molecular Plant, 2019, 12(8): 1157-1166. |
| [13] | GAUR V S, SOOD S, GUZMÁN C, et al. Molecular insights on the origin and development of waxy genotypes in major crop plants[J]. Briefings in Functional Genomics, 2024, 23(3): 193-213. |
| [14] | RENGASAMY B, MANNA M, THAJUDDIN N B, et al. Breeding rice for yield improvement through CRISPR/Cas9 genome editing method: current technologies and examples[J]. Physiology and Molecular Biology of Plants, 2024, 30(2): 185-198. |
| [15] | 李可, 吴传银, 隋毅. CRISPR/Cas基因编辑技术在水稻育种中的研究进展[J]. 科学通报, 2025, 70(16): 2483-2494. |
| LI K, WU C Y, SUI Y. Research progress of CRISPR/Cas gene editing technology in rice breeding[J]. Chinese Science Bulletin, 2025, 70(16): 2483-2494. | |
| [16] | OKPALA N E, MO Z W, DUAN M Y, et al. The genetics and biosynthesis of 2-acetyl-1-pyrroline in fragrant rice[J]. Plant Physiology and Biochemistry, 2019, 135: 272-276. |
| [17] | 季新, 李猛, 杨阳, 等. 基于CRISPR/Cas9技术创制宜直播香型水稻[J]. 分子植物育种, 2023, 21(19): 6381-6389. |
| JI X, LI M, YANG Y, et al. Production of adapted direct-seeding fragrant rice based on CRISPR/Cas9 technology[J]. Molecular Plant Breeding, 2023, 21(19): 6381-6389. | |
| [18] | 胡彬华, 蒲志刚, 何志渊, 等. 利用CRISPR/Cas9技术敲除OsNramp5创制镉低积累水稻新种质[J]. 植物遗传资源学报, 2024, 25(7): 1211-1219. |
| HU B H, PU Z G, HE Z Y, et al. Generating low cadmium accumulation new rice germplasms by editing OsNramp5 using CRISPR/Cas9 technology[J]. Journal of Plant Genetic Resources, 2024, 25(7): 1211-1219. | |
| [19] | SASAKI A, YAMAJI N, YOKOSHO K, et al. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice[J]. The Plant Cell, 2012, 24(5): 2155-2167. |
| [20] | 赵考诚, 黄玲玲, 余志平, 等. 近20年浙江省糯稻品种育种现状及发展策略[J]. 中国稻米, 2024, 30(3): 84-87. |
| ZHAO K C, HUANG L L, YU Z P, et al. Breeding status and development strategies of glutinous rice varieties in Zhejiang Province in the past 20 years[J]. China Rice, 2024, 30(3): 84-87. | |
| [21] | 丁正权, 来乐春, 王士磊, 等. 优质食味长粒粳稻嘉禾香1号[J]. 中国种业, 2023(2): 126-127. |
| DING Z Q, LAI L C, WANG S L, et al. Jiahexiang1, a long-grain Japonica rice variety with fine-eating quality[J]. China Seed Industry, 2023(2): 126-127. | |
| [22] | MANSUETO L, FUENTES R R, BORJA F N, et al. Rice SNP-seek database update: new SNPs, indels, and queries[J]. Nucleic Acids Research, 2017, 45(D1): D1075-D1081. |
| [23] | JIANG C H, RASHID M A R, ZHANG Y H, et al. Genome wide association study on development and evolution of glutinous rice[J]. BMC Genomic Data, 2022, 23(1): 33. |
| [24] | BRADBURY P J, ZHANG Z W, KROON D E, et al. TASSEL: software for association mapping of complex traits in diverse samples[J]. Bioinformatics, 2007, 23(19): 2633-2635. |
| [25] | LIU W Z, XIE X R, MA X L, et al. DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations[J]. Molecular Plant, 2015, 8(9): 1431-1433. |
| [26] | HIRANO H Y, EIGUCHI M, SANO Y. A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice[J]. Molecular Biology and Evolution, 1998, 15(8): 978-987. |
| [27] | 汪秉琨, 张慧, 洪汝科, 等. CRISPR/Cas9系统编辑水稻Wx基因[J]. 中国水稻科学, 2018, 32(1): 35-42. |
| WANG B K, ZHANG H, HONG R K, et al. Wx gene editing via CRISPR/Cas9 system in rice[J]. Chinese Journal of Rice Science, 2018, 32(1): 35-42. | |
| [28] | 丁正权, 潘月云, 施扬, 等. 基于基因芯片的嘉禾系列长粒优质食味粳稻综合评价与比较[J]. 中国水稻科学, 2024, 38(4): 397-408. |
| DING Z Q, PAN Y Y, SHI Y, et al. Comprehensive evaluation and comparative analysis of Jiahe series long-grain japonica rice with high eating quality based on gene chip technology[J]. Chinese Journal of Rice Science, 2024, 38(4): 397-408. | |
| [29] | HUANG L C, LI Q F, ZHANG C Q, et al. Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system[J]. Plant Biotechnology Journal, 2020, 18(11): 2164-2166. |
| [30] | XU Y, LIN Q P, LI X F, et al. Fine-tuning the amylose content of rice by precise base editing of the Wx gene[J]. Plant Biotechnology Journal, 2021, 19(1): 11-13. |
| [31] | ZENG D C, LIU T L, MA X L, et al. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5'UTR-intron editing improves grain quality in rice[J]. Plant Biotechnology Journal, 2020, 18(12): 2385-2387. |
| [32] | 周苏淮, 李芳, 李娟, 等. 植物基因组精准编辑技术: 引导编辑[J]. 科学通报, 2025, 70(16): 2414-2422. |
| ZHOU S H, LI F, LI J, et al. Plant genome precise editing technology-prime editing[J]. Chinese Science Bulletin, 2025, 70(16): 2414-2422. |
| [1] | 朱长松, 纳琦婷, 张梦卓, 曹慧, 刘诗颖, 张正科, 孟兰环. SlCHRC基因对高温环境下番茄花耐热性的影响[J]. 浙江农业学报, 2026, 38(1): 67-75. |
| [2] | 裴惠民, 巫明明, 翟荣荣, 叶靖, 金月, 朱仪, 侯建军, 朱国富, 叶胜海. 低镉水稻基因功能与新品种培育研究进展[J]. 浙江农业学报, 2025, 37(9): 2012-2020. |
| [3] | 谭诗逸, 俞国红, 薛向磊, 赵颖雷, 许宝玉, 张成浩. 工厂化水稻育秧盘搬运装置设计与试验[J]. 浙江农业学报, 2025, 37(7): 1545-1555. |
| [4] | 张智, 何豪豪, 郁妙, 许剑锋. 化肥减量配施土壤改良剂对土壤酸度、土壤养分和水稻产量的影响[J]. 浙江农业学报, 2025, 37(6): 1301-1308. |
| [5] | 林小兵, 黎江, 成艳红, 王斌强, 何绍浪, 黄尚书, 黄欠如. 不同有机物料对土壤微生物生物量、矿质氮含量与水稻产量的影响[J]. 浙江农业学报, 2025, 37(6): 1309-1318. |
| [6] | 苏扬, 商小兰, 钱忠明, 吴林根, 黄佳琦, 庄海峰, 赵宇飞, 党洪阳, 徐立军. 腐熟剂与生物炭协同强化秸秆还田对土壤质量和水稻生长的影响[J]. 浙江农业学报, 2025, 37(5): 1139-1148. |
| [7] | 应永飞, 韩东轩, 孟芳, 俞遴, 沈佳栾, 汪开英. 沼液替代化肥对水稻产量、品质和土壤特性的影响[J]. 浙江农业学报, 2025, 37(4): 880-891. |
| [8] | 宋欣录, 范书红, 武桄旗, 展梦琪, 侯倩, 李明月, 徐艳. 铜-菲复合污染对分蘖期水稻根系生理特性和污染物积累的影响[J]. 浙江农业学报, 2025, 37(3): 521-529. |
| [9] | 雷志伟, 李新欣, 徐恒, 张恒, 朱英, 张华. 利用染色体片段替换系鉴定水稻二化螟抗性QTL[J]. 浙江农业学报, 2025, 37(3): 530-537. |
| [10] | 谢昶琰, 金雨濛, 张苗, 董青君, 李青, 纪力, 钟平, 陈川, 章安康. 利用河道淤泥开发机插水稻秧苗营养土及其应用效果[J]. 浙江农业学报, 2025, 37(3): 538-547. |
| [11] | 兰雪成, 赵凤亮, 张光旭, 李杨, 郭晓红. 纳米氧化锌和纳米氧化硅对水稻种子萌发的影响[J]. 浙江农业学报, 2025, 37(2): 269-277. |
| [12] | 李建强, 魏倩倩, 刘晓霞, 张均华, 朱春权. 优化施肥措施对水稻产量和土壤养分平衡的影响[J]. 浙江农业学报, 2025, 37(2): 438-446. |
| [13] | 郝中娜, 邱海萍, 柴荣耀, 韩展誉, 张震, 王艳丽, 王教瑜, 刘鑫. 浙江省2014—2023年区域试验中水稻品种的稻瘟病抗性分析[J]. 浙江农业学报, 2025, 37(12): 2449-2457. |
| [14] | 杨锦皓, 江洁, 刘行, 李福强. 基于离散元法的水稻种子参数标定与旱作排种仿真[J]. 浙江农业学报, 2025, 37(11): 2376-2386. |
| [15] | 李新欣, 徐恒, 宋涛, 袁熹, 孙梅好, 朱英, 张华. 水稻耐高温遗传基础和调控机制的研究进展[J]. 浙江农业学报, 2025, 37(11): 2426-2440. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||