Acta Agriculturae Zhejiangensis ›› 2025, Vol. 37 ›› Issue (10): 2104-2115.DOI: 10.3969/j.issn.1004-1524.20250023
• Horticultural Science • Previous Articles Next Articles
Identification of the HKT gene family members in Populus species and analysis of their expression patterns under salt stress
LIAO Xiaolong1( ), WANG Xingsheng1, CHEN Yong1, LI Bin1, HONG Sidan2, MEI Lina2, GUO Ying2,*(
), WANG Xingsheng1, CHEN Yong1, LI Bin1, HONG Sidan2, MEI Lina2, GUO Ying2,*( )
)
- 1. Ili Experimental Center of Tree Breeding for Improved Varieties(Plain Forestry Farm), Ili Kazak Autonomous Prefecture, Ili 835311, Xinjiang, China
2. Jiangsu Key Laboratory for Poplar Germplasm Enhancement and Variety Improvement, Co-Innovation Center for the Sustainable Forestry in Southern China, College of Forestry and Grassland, College of Soil and Water Conservation, Nanjing Forestry University, Nanjing 210037, China
-
Received:2025-01-09Online:2025-10-25Published:2025-11-13
CLC Number:
Cite this article
LIAO Xiaolong, WANG Xingsheng, CHEN Yong, LI Bin, HONG Sidan, MEI Lina, GUO Ying. Identification of the HKT gene family members in Populus species and analysis of their expression patterns under salt stress[J]. Acta Agriculturae Zhejiangensis, 2025, 37(10): 2104-2115.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.zjnyxb.cn/EN/10.3969/j.issn.1004-1524.20250023
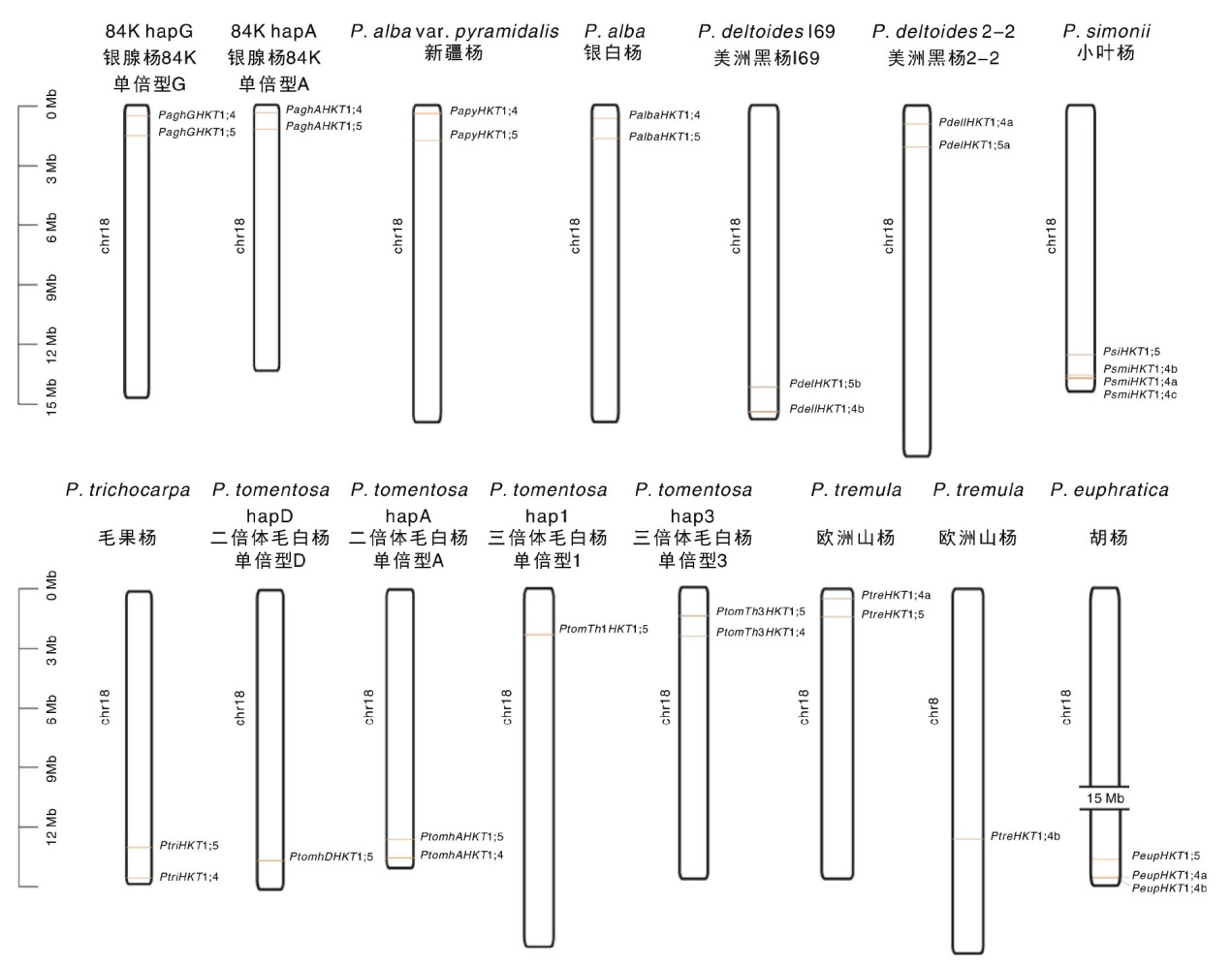
| 种/品种 Species/Cultivar | 单倍型 Haplotype | 组装方式 Assembly strategy |
|---|---|---|
| 欧洲山杨P. tremula | / | 基于参考基因组的基因组组装 |
| 毛果杨P. trichocarpa | / | Reference based assembly |
| 胡杨P. euphratica | / | 从头组装De novo assembly |
| 小叶杨P. simonii | / | |
| 银白杨P. alba | / | |
| 新疆杨P. alba var. pyramidalis | / | |
| 美洲黑杨I69 P. deltoides I69 | / | |
| 美洲黑杨2-2 P. deltoides 2-2 | / | |
| 银腺杨(银腺杨84K)P. alba×P. tremula var. glandulosa clone 84K | 单倍型A hapA | |
| 单倍型G hapG | ||
| 二倍体毛白杨P. tomentosa | 单倍型A hapA | |
| 单倍型D hapD | ||
| 三倍体毛白杨P.×tomentosa Carr. clone 741 | 单倍型1 hap1 | |
| 单倍型2 hap2 | ||
| 单倍型3 hap3 |
Table 1 Statistical table of Populus plants genome information
| 种/品种 Species/Cultivar | 单倍型 Haplotype | 组装方式 Assembly strategy |
|---|---|---|
| 欧洲山杨P. tremula | / | 基于参考基因组的基因组组装 |
| 毛果杨P. trichocarpa | / | Reference based assembly |
| 胡杨P. euphratica | / | 从头组装De novo assembly |
| 小叶杨P. simonii | / | |
| 银白杨P. alba | / | |
| 新疆杨P. alba var. pyramidalis | / | |
| 美洲黑杨I69 P. deltoides I69 | / | |
| 美洲黑杨2-2 P. deltoides 2-2 | / | |
| 银腺杨(银腺杨84K)P. alba×P. tremula var. glandulosa clone 84K | 单倍型A hapA | |
| 单倍型G hapG | ||
| 二倍体毛白杨P. tomentosa | 单倍型A hapA | |
| 单倍型D hapD | ||
| 三倍体毛白杨P.×tomentosa Carr. clone 741 | 单倍型1 hap1 | |
| 单倍型2 hap2 | ||
| 单倍型3 hap3 |
| 基因名 Gene name | 正向引物序列 Forward primer sequence(5'-3') | 反向引物序列 Reverse primer sequence(5'-3') |
|---|---|---|
| PaghHKT1;5 | GGTGCTCTTCGTGGTTAT | CACAAAGATGGCTAAGGT |
| PaghAHKT1;4 | GCCAGCATTCTTCTTATT | TGACCACAAACAGCACCA |
| PaghGHKT1;4 | TCCCAACTCATTGTCTTC | CAAACAGCACCAAGATAG |
| UBQ | GTTGATTTTTGCTGGGAAGC | GATCTTGGCCTTCACGTTGT |
Table 2 Primer sequences for qRT-PCR
| 基因名 Gene name | 正向引物序列 Forward primer sequence(5'-3') | 反向引物序列 Reverse primer sequence(5'-3') |
|---|---|---|
| PaghHKT1;5 | GGTGCTCTTCGTGGTTAT | CACAAAGATGGCTAAGGT |
| PaghAHKT1;4 | GCCAGCATTCTTCTTATT | TGACCACAAACAGCACCA |
| PaghGHKT1;4 | TCCCAACTCATTGTCTTC | CAAACAGCACCAAGATAG |
| UBQ | GTTGATTTTTGCTGGGAAGC | GATCTTGGCCTTCACGTTGT |
| 蛋白质ID Protein ID | 基因ID Gene ID | 氨基酸长度 Amino acid length/aa | 分子量 Molecular weight/ku | 等电点 Isoelectric point | |||
|---|---|---|---|---|---|---|---|
| PaghAHKT1;5 | Pop_A18G018657 | 535 | 60.02 | 9.41 | |||
| PaghAHKT1;4 | Pop_A18G034182 | 470 | 52.95 | 9.01 | |||
| PaghGHKT1;5 | Pop_G18G019156 | 473 | 52.68 | 9.22 | |||
| PaghGHKT1;4 | Pop_G18G086088 | 206 | 23.22 | 9.49 | |||
| PalbaHKT1;5 | XP_034918867.1 | 535 | 60.02 | 9.41 | |||
| PalbaHKT1;4 | XP_034906209.1 | 521 | 58.94 | 9.02 | |||
| PapyHKT1;4 | GWHPACDA010899 | 621 | 69.63 | 9.03 | |||
| PapyHKT1;5 | GWHPACDA011033 | 451 | 50.37 | 9.04 | |||
| PtomTh1HKT1;5 | GWHPBJCQ043518 | 535 | 59.98 | 9.33 | |||
| PtomTh3HKT1;5 | GWHPBJCQ045429 | 535 | 59.79 | 9.26 | |||
| PtomTh3HKT1;4 | GWHPBJCQ045526 | 501 | 56.69 | 9.16 | |||
| PdelHKT1;5b | EVM0018125.1 | 516 | 58.11 | 9.51 | |||
| PdelHKT1;4b | EVM0019360.1 | 238 | 27.38 | 8.64 | |||
| PdelHKT1;5a | KAH8481580.1 | 473 | 52.73 | 9.27 | |||
| PdelHKT1;4a | KAH8481435.1 | 166 | 18.90 | 9.24 | |||
| PeupHKT1;5 | GWHPAAYU005564.1 | 535 | 59.96 | 9.29 | |||
| PeupHKT1;4b | GWHPAAYU005664.1 | 521 | 58.86 | 9.13 | |||
| PeupHKT1;4a | GWHPAAYU005662.1 | 422 | 47.86 | 8.85 | |||
| PsiHKT1;5 | Simonii00009649-RA | 535 | 59.94 | 9.39 | |||
| PsiHKT1;4c | Simonii00039973-RA | 521 | 58.81 | 9.15 | |||
| PsiHKT1;4b | Simonii00039952-RA | 521 | 59.00 | 9.13 | |||
| PsiHKT1;4a | Simonii00039964-RA | 498 | 56.32 | 9.07 | |||
| PtomhAHKT1;5 | KAG6741040.1 | 535 | 60.04 | 9.41 | |||
| PtomhAHKT1;4 | KAG6741120.1 | 500 | 56.53 | 9.16 | |||
| PtomhDHKT1;5 | KAG6740023.1 | 473 | 52.60 | 9.16 | |||
| PtreHKT1;5 | Potra2n18c32115.1 | 535 | 59.79 | 9.45 | |||
| PtreHKT1;4b | Potra2n8c18208.1 | 521 | 59.10 | 9.27 | |||
| PtreHKT1;4a | Potra2n18c32004.1 | 397 | 44.62 | 9.43 | |||
| PtriHKT1;5 | Potri.018G132200.1.p | 535 | 60.09 | 9.45 | |||
| PtriHKT1;4 | Potri.018G147501.2.p | 168 | 19.40 | 9.62 | |||
Table 3 Physicochemical properties of HKT protein in Populus plants
| 蛋白质ID Protein ID | 基因ID Gene ID | 氨基酸长度 Amino acid length/aa | 分子量 Molecular weight/ku | 等电点 Isoelectric point | |||
|---|---|---|---|---|---|---|---|
| PaghAHKT1;5 | Pop_A18G018657 | 535 | 60.02 | 9.41 | |||
| PaghAHKT1;4 | Pop_A18G034182 | 470 | 52.95 | 9.01 | |||
| PaghGHKT1;5 | Pop_G18G019156 | 473 | 52.68 | 9.22 | |||
| PaghGHKT1;4 | Pop_G18G086088 | 206 | 23.22 | 9.49 | |||
| PalbaHKT1;5 | XP_034918867.1 | 535 | 60.02 | 9.41 | |||
| PalbaHKT1;4 | XP_034906209.1 | 521 | 58.94 | 9.02 | |||
| PapyHKT1;4 | GWHPACDA010899 | 621 | 69.63 | 9.03 | |||
| PapyHKT1;5 | GWHPACDA011033 | 451 | 50.37 | 9.04 | |||
| PtomTh1HKT1;5 | GWHPBJCQ043518 | 535 | 59.98 | 9.33 | |||
| PtomTh3HKT1;5 | GWHPBJCQ045429 | 535 | 59.79 | 9.26 | |||
| PtomTh3HKT1;4 | GWHPBJCQ045526 | 501 | 56.69 | 9.16 | |||
| PdelHKT1;5b | EVM0018125.1 | 516 | 58.11 | 9.51 | |||
| PdelHKT1;4b | EVM0019360.1 | 238 | 27.38 | 8.64 | |||
| PdelHKT1;5a | KAH8481580.1 | 473 | 52.73 | 9.27 | |||
| PdelHKT1;4a | KAH8481435.1 | 166 | 18.90 | 9.24 | |||
| PeupHKT1;5 | GWHPAAYU005564.1 | 535 | 59.96 | 9.29 | |||
| PeupHKT1;4b | GWHPAAYU005664.1 | 521 | 58.86 | 9.13 | |||
| PeupHKT1;4a | GWHPAAYU005662.1 | 422 | 47.86 | 8.85 | |||
| PsiHKT1;5 | Simonii00009649-RA | 535 | 59.94 | 9.39 | |||
| PsiHKT1;4c | Simonii00039973-RA | 521 | 58.81 | 9.15 | |||
| PsiHKT1;4b | Simonii00039952-RA | 521 | 59.00 | 9.13 | |||
| PsiHKT1;4a | Simonii00039964-RA | 498 | 56.32 | 9.07 | |||
| PtomhAHKT1;5 | KAG6741040.1 | 535 | 60.04 | 9.41 | |||
| PtomhAHKT1;4 | KAG6741120.1 | 500 | 56.53 | 9.16 | |||
| PtomhDHKT1;5 | KAG6740023.1 | 473 | 52.60 | 9.16 | |||
| PtreHKT1;5 | Potra2n18c32115.1 | 535 | 59.79 | 9.45 | |||
| PtreHKT1;4b | Potra2n8c18208.1 | 521 | 59.10 | 9.27 | |||
| PtreHKT1;4a | Potra2n18c32004.1 | 397 | 44.62 | 9.43 | |||
| PtriHKT1;5 | Potri.018G132200.1.p | 535 | 60.09 | 9.45 | |||
| PtriHKT1;4 | Potri.018G147501.2.p | 168 | 19.40 | 9.62 | |||
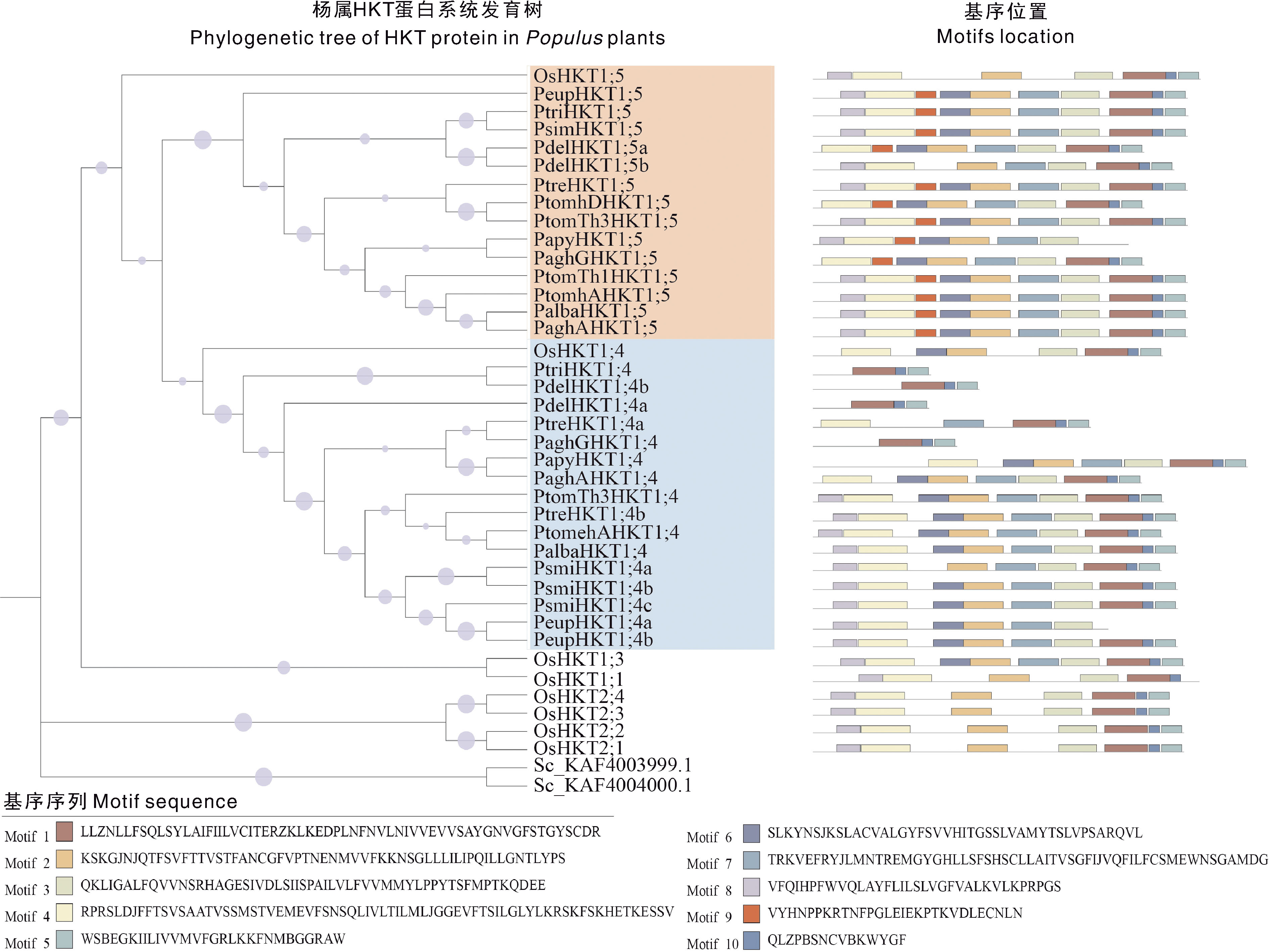
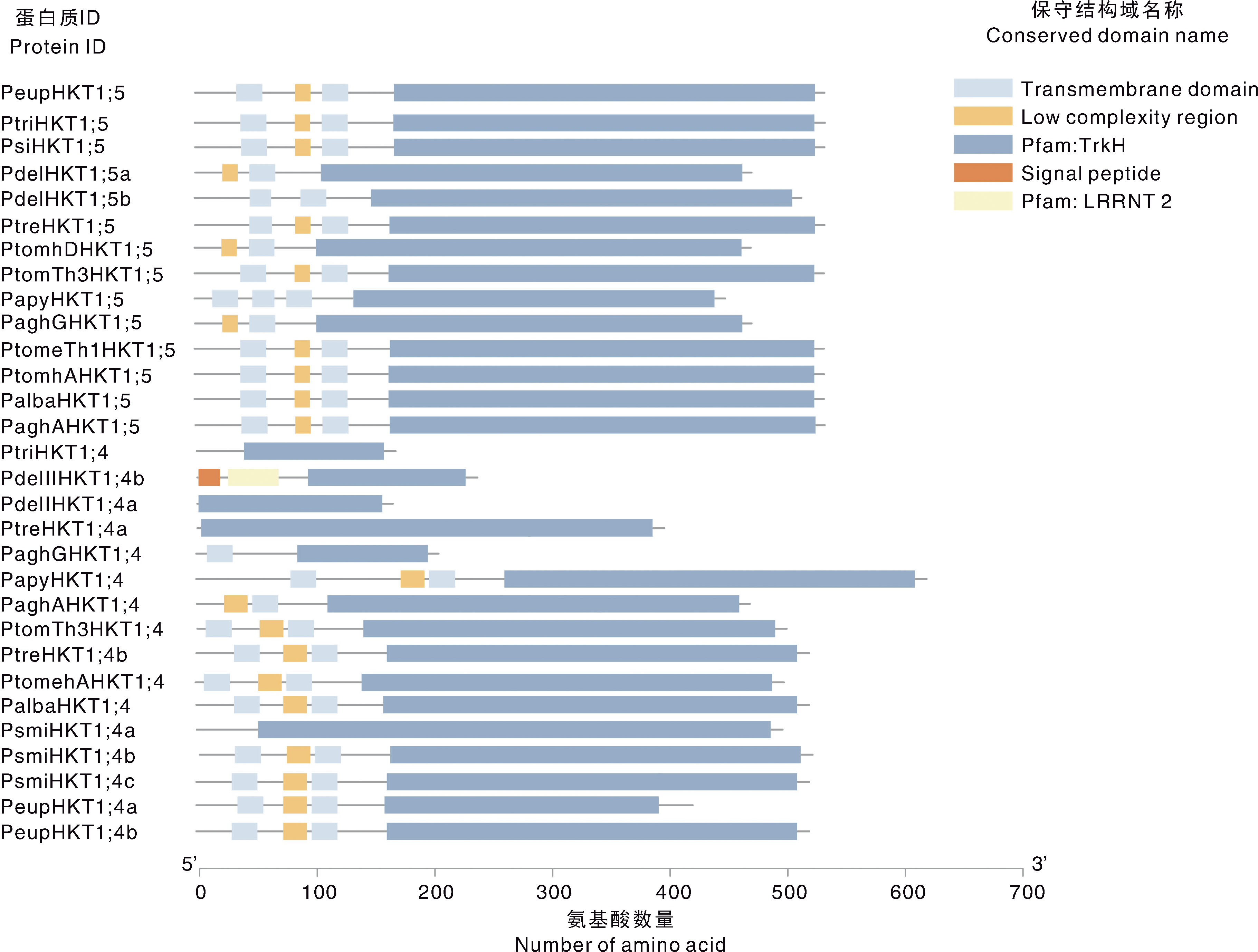
Fig.2 Phylogenetic tree and conserved motifs of HKT gene family members in Populus plants The purple dots indicate the nodes of the bootstrap between 0.315-1.000. Different coloured cubes represent different conserved motifs.
| 种/品种 Species/Cultivar | 单倍型 Haplotype | 同源组1 OG01 | 同源组2 OG02 | 同源组3 OG03 | 同源组4 OG04 | 同源组5 OG05 |
|---|---|---|---|---|---|---|
| 银腺杨84K P. alba×P. tremula var. glandulosa clone 84K | 单倍型A hapA | 1 | 1 | 0 | 0 | 0 |
| 单倍型G hapG | 1 | 1 | 0 | 0 | 0 | |
| 银白杨P. alba | / | 0 | 0 | 2 | 0 | 0 |
| 新疆杨P. alba var. pyramidalis | / | 1 | 1 | 0 | 0 | 0 |
| 三倍体毛白杨P.×tomentosa Carr. clone 741 | 单倍型1 hap 1 | 0 | 0 | 0 | 1 | 0 |
| 三倍体毛白杨P.×tomentosa Carr. clone 741 | 单倍型3 hap 3 | 0 | 0 | 2 | 0 | 0 |
| 美洲黑杨2-2 P. deltoides 2-2 | / | 1 | 1 | 0 | 0 | 0 |
| 美洲黑杨Ⅰ69 P. deltoides I69 | / | 1 | 1 | 0 | 0 | 0 |
| 胡杨P. euphratica | / | 2 | 1 | 0 | 0 | 0 |
| 小叶杨P. simonii | / | 3 | 1 | 0 | 0 | 0 |
| 二倍体毛白杨P. tomentosa | 单倍型A hap A | 1 | 1 | 0 | 0 | 0 |
| 二倍体毛白杨P. tomentosa | 单倍型D hap D | 0 | 0 | 0 | 1 | 0 |
| 欧洲山杨P. tremula | / | 2 | 1 | 0 | 0 | 0 |
| 毛果杨P. trichocarpa | / | 1 | 1 | 0 | 0 | 0 |
| 水稻Oryza sativa | / | 0 | 0 | 1 | 3 | 4 |
| 总计Total | / | 14 | 10 | 5 | 5 | 4 |
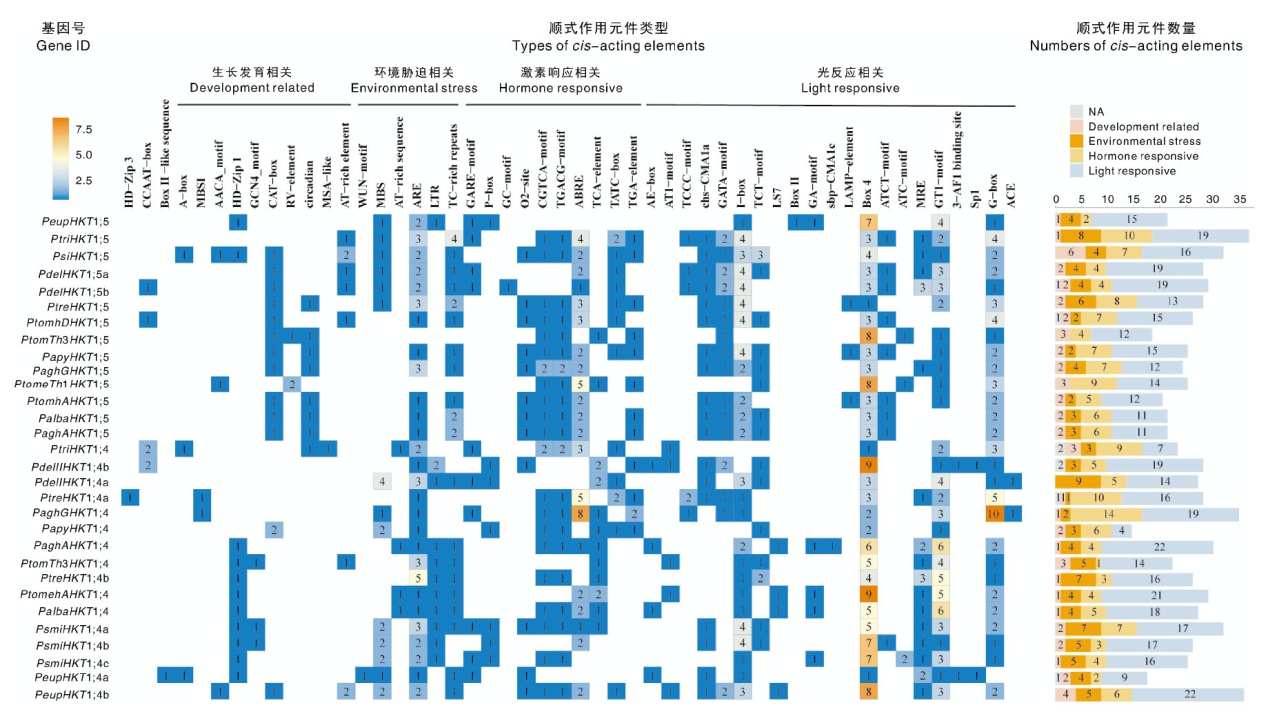
Table 4 Homology of HKT family members in Populus plants
| 种/品种 Species/Cultivar | 单倍型 Haplotype | 同源组1 OG01 | 同源组2 OG02 | 同源组3 OG03 | 同源组4 OG04 | 同源组5 OG05 |
|---|---|---|---|---|---|---|
| 银腺杨84K P. alba×P. tremula var. glandulosa clone 84K | 单倍型A hapA | 1 | 1 | 0 | 0 | 0 |
| 单倍型G hapG | 1 | 1 | 0 | 0 | 0 | |
| 银白杨P. alba | / | 0 | 0 | 2 | 0 | 0 |
| 新疆杨P. alba var. pyramidalis | / | 1 | 1 | 0 | 0 | 0 |
| 三倍体毛白杨P.×tomentosa Carr. clone 741 | 单倍型1 hap 1 | 0 | 0 | 0 | 1 | 0 |
| 三倍体毛白杨P.×tomentosa Carr. clone 741 | 单倍型3 hap 3 | 0 | 0 | 2 | 0 | 0 |
| 美洲黑杨2-2 P. deltoides 2-2 | / | 1 | 1 | 0 | 0 | 0 |
| 美洲黑杨Ⅰ69 P. deltoides I69 | / | 1 | 1 | 0 | 0 | 0 |
| 胡杨P. euphratica | / | 2 | 1 | 0 | 0 | 0 |
| 小叶杨P. simonii | / | 3 | 1 | 0 | 0 | 0 |
| 二倍体毛白杨P. tomentosa | 单倍型A hap A | 1 | 1 | 0 | 0 | 0 |
| 二倍体毛白杨P. tomentosa | 单倍型D hap D | 0 | 0 | 0 | 1 | 0 |
| 欧洲山杨P. tremula | / | 2 | 1 | 0 | 0 | 0 |
| 毛果杨P. trichocarpa | / | 1 | 1 | 0 | 0 | 0 |
| 水稻Oryza sativa | / | 0 | 0 | 1 | 3 | 4 |
| 总计Total | / | 14 | 10 | 5 | 5 | 4 |
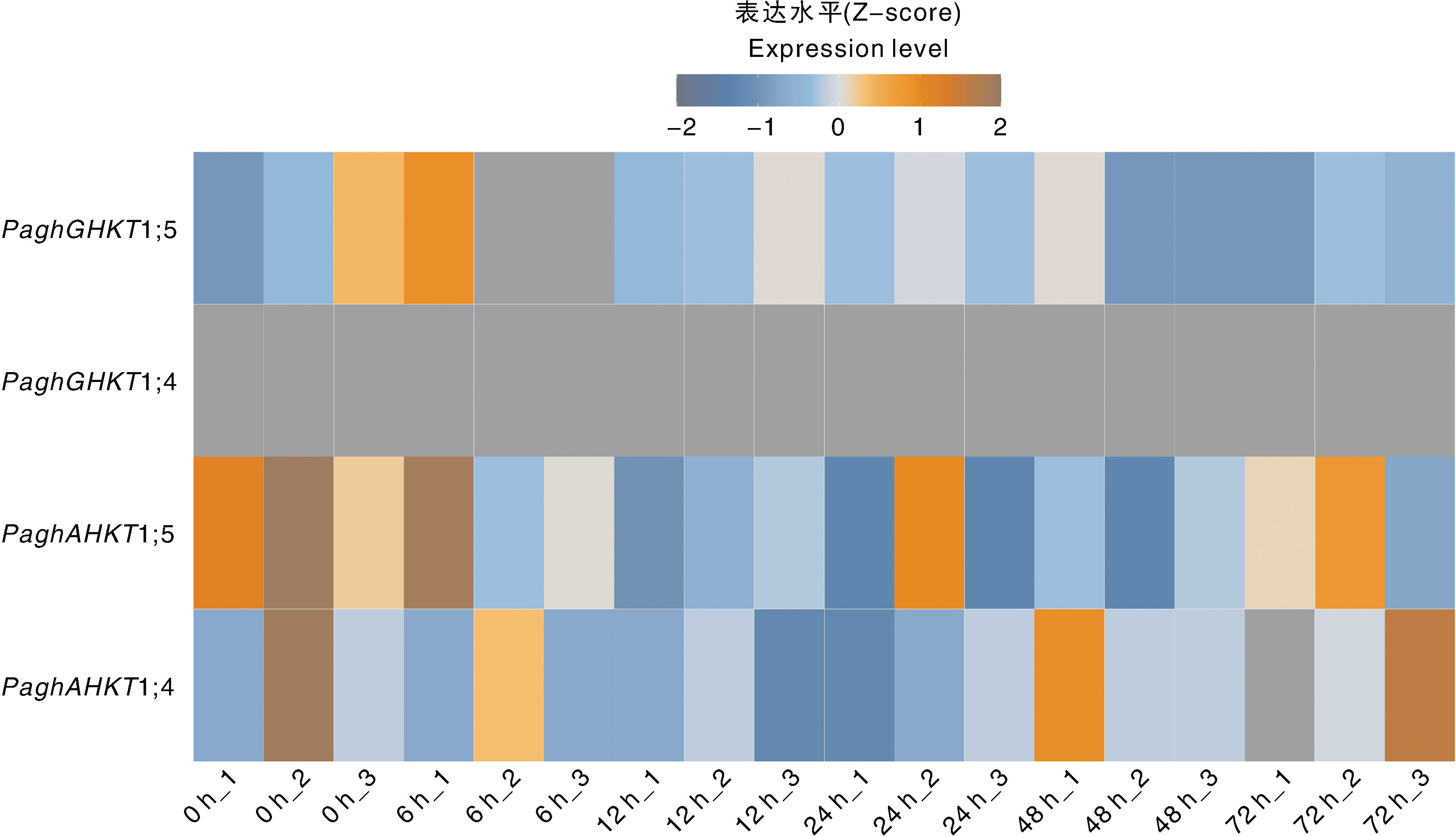
Fig.5 Expression level of HKT genes in the leaves of poplar clone 84K under 100 mmol·L-1 NaCl treatment The labels 1, 2, and 3 below the x-axis indicate the three biological replicates for each time point(0, 6, 12, 24, 48, and 72 h) under 100 mmol·L 1 NaCl treatment. The expression levels were normalized using the Z-score method.
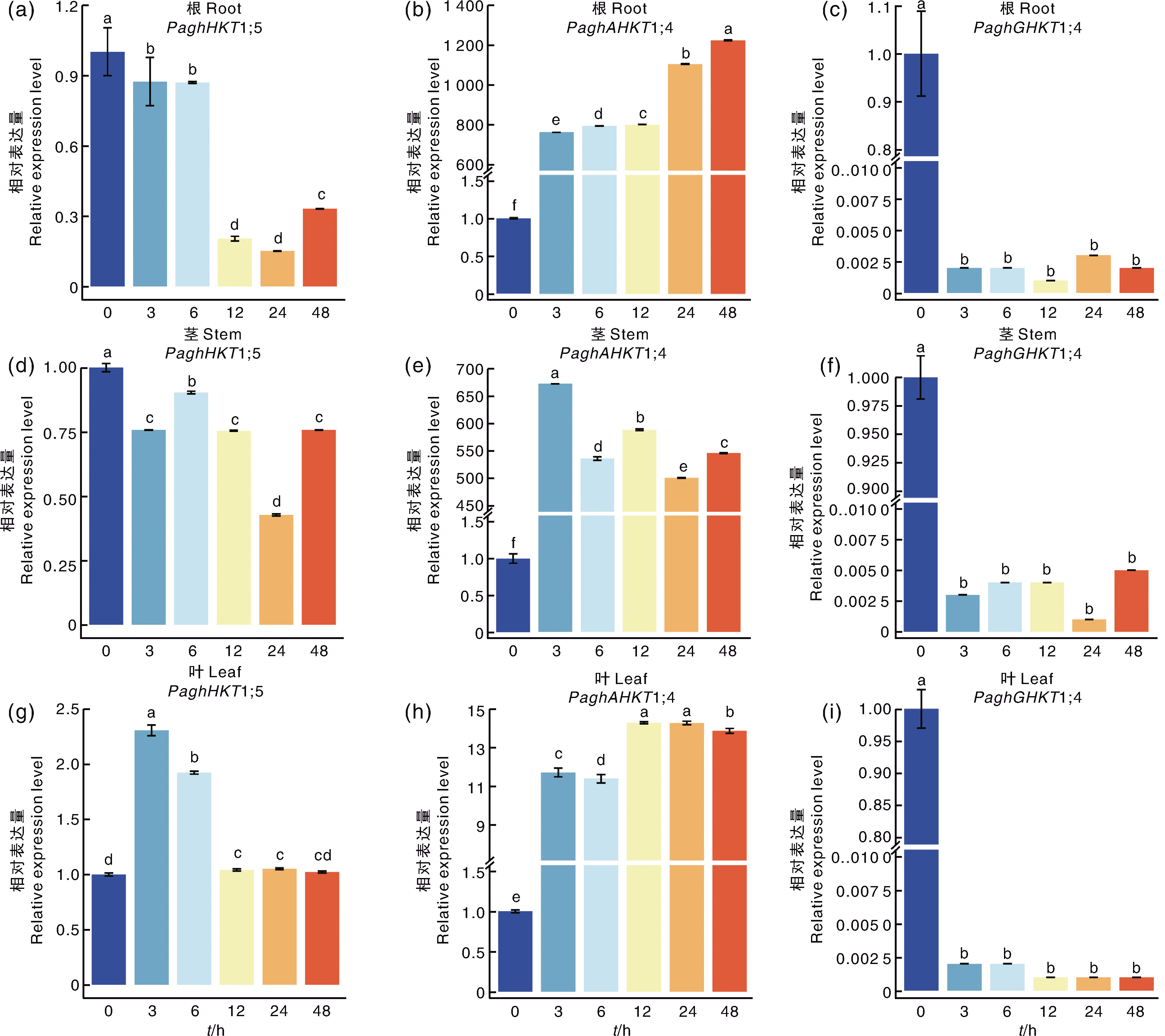
Fig.6 Expression level of HKT genes in different tissues in poplar clone 84K under 150 mmol·L-1 NaCl treatment Data marked without the same lowercase letter in each column indicated significant differences at p<0.05.
| [1] | 欧阳易佳. 唤醒“沉睡”耕地资源让盐碱沙荒变“大国粮仓”[EB/OL]. (2024-01-13)[2024-06-30]. https://news.sina.com.cn/zx/gj/2024-01-14/doc-inacmkcr9735840.shtml. |
| [2] | 杜建辉. 重度盐碱化耕地治理的N种新模式[EB/OL]. (2023-11-15)[2024-06-30]. https://www.btzx.com.cn/web/2023/11/15/ARTI1700035965497340.html. |
| [3] | 梅隆, 刘自艰, 赵倩倩. 从“治理”到“适应”,重新认识盐碱地的价值[EB/OL]. (2022-07-29)[2024-06-30]. https://szb.farmer.com.cn/2022/20220729/20220729_008/20220729_008_1.htm. |
| [4] | 刘钰, 张艳华, 方升佐. 株行距配置和无性系对杨树人工林碳储量的影响[J]. 森林与环境学报, 2024, 44(3): 242-249. |
| LIU Y, ZHANG Y H, FANG S Z. Effects of planting spacing configurations and clones on carbon storage in poplar plantations[J]. Journal of Forest and Environment, 2024, 44(3): 242-249. (in Chinese with English abstract) | |
| [5] | 姚诗雨, 王杰, 黄文娟, 等. 不同展叶物候期胡杨离子分布、吸收和运输特征及其与土壤盐分关系[J]. 西北植物学报, 2023, 43(12): 2118-2129. |
| YAO S Y, WANG J, HUANG W J, et al. Distribution, uptake and transport characteristics of Populus euphratica ions at different leaf phenological stages and their relationship with soil salinity[J]. Acta Botanica Boreali-Occidentalia Sinica, 2023, 43(12): 2118-2129. (in Chinese with English abstract) | |
| [6] | 左照江, 张汝民, 高岩. 盐胁迫下植物细胞离子流变化的研究进展[J]. 浙江农林大学学报, 2014, 31(5): 805-811. |
| ZUO Z J, ZHANG R M, GAO Y. Advances in plant cell ion flux with salt stress: a review[J]. Journal of Zhejiang A&F University, 2014, 31(5): 805-811. (in Chinese with English abstract) | |
| [7] | SHABALA S. Ionic and osmotic components of salt stress specifically modulate net ion fluxes from bean leaf mesophyll[J]. Plant, Cell & Environment, 2000, 23(8): 825-837. |
| [8] | SHI H Z, LEE B H, WU S J, et al. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana[J]. Nature Biotechnology, 2002, 21(1): 81-85. |
| [9] | MUNNS R, TESTER M. Mechanisms of salinity tolerance[J]. Annual Review of Plant Biology, 2008, 59: 651-681. |
| [10] | VERA-ESTRELLA R, BARKLA B J, BOHNERT H J, et al. Salt stress in Mesembryanthemum crystallinum L. cell suspensions activates adaptive mechanisms similar to those observed in the whole plant[J]. Planta, 1999, 207(3): 426-435. |
| [11] | BEILBY M J, SHEPHERD V A. Modeling the current-voltage characteristics of charophyte membranes. II. the effect of salinity on membranes of Lamprothamnium papulosum[J]. The Journal of Membrane Biology, 2001, 181(2): 77-89. |
| [12] | SHABALA L, CUIN T A, NEWMAN I A, et al. Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants[J]. Planta, 2005, 222(6): 1041-1050. |
| [13] | RIEDELSBERGER J, MILLER J K, VALDEBENITO-MATURANA B, et al. Plant HKT channels: an updated view on structure, function and gene regulation[J]. International Journal of Molecular Sciences, 2021, 22(4): 1892. |
| [14] | BYRT C S, XU B, KRISHNAN M, et al. The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat[J]. The Plant Journal, 2014, 80(3): 516-526. |
| [15] | HAMAMOTO S, HORIE T, HAUSER F, et al. HKT transporters mediate salt stress resistance in plants: from structure and function to the field[J]. Current Opinion in Biotechnology, 2015, 32: 113-120. |
| [16] | XIAO L Y, SHI Y Y, WANG R, et al. The transcription factor OsMYBc and an E3 ligase regulate expression of a K+ transporter during salt stress[J]. Plant Physiology, 2022, 190(1): 843-859. |
| [17] | HUA Y P, PEI M N, SONG H L, et al. Boron confers salt tolerance through facilitating BnaA2.HKT1-mediated root xylem Na+ unloading in rapeseed (Brassica napus L.)[J]. The Plant Journal, 2024, 120(4): 1326-1342. |
| [18] | UCHIYAMA T, SAITO S, YAMANASHI T, et al. The HKT1 Na+ transporter protects plant fertility by decreasing Na+ content in stamen filaments[J]. Science Advances, 2023, 9(22): eadg5495. |
| [19] | 徐文君, 刘兆普, 隆小华, 等. 农杆菌介导转AtNHX1基因杨树的获得[J]. 植物生理学通讯, 2007, 43(3): 413-416. |
| XU W J, LIU Z P, LONG X H, et al. Transformation of populus x euramericana with AtNHX1 gene mediated by Agrobacterium tumefaciens[J]. Plant Physiology Communications, 2007, 43(3): 413-416. (in Chinese with English abstract) | |
| [20] | SUN J, CHEN S L, DAI S X, et al. NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species[J]. Plant Physiology, 2009, 149(2): 1141-1153. |
| [21] | ZHANG Z Y, CHEN Y, ZHANG J L, et al. Improved genome assembly provides new insights into genome evolution in a desert poplar (Populus euphratica)[J]. Molecular Ecology Resources, 2020, 20(3): 781-794. |
| [22] | LIU Y J, WANG X R, ZENG Q Y. De novo assembly of white poplar genome and genetic diversity of white poplar population in Irtysh River basin in China[J]. Science China Life Sciences, 2019, 62(5): 609-618. |
| [23] | ZHANG L, ZHAO J T, BI H, et al. Bioinformatic analysis of chromatin organization and biased expression of duplicated genes between two poplars with a common whole-genome duplication[J]. Horticulture Research, 2021, 8: 62. |
| [24] | WU H N, YAO D, CHEN Y H, et al. De novo genome assembly of Populus simonii further supports that Populus simonii and Populus trichocarpa belong to different sections[J]. G3 Genes Genomes Genetics, 2020, 10(2): 455-466. |
| [25] | WANG J, DING J H, TAN B Y, et al. A major locus controls local adaptation and adaptive life history variation in a perennial plant[J]. Genome Biology, 2018, 19(1): 72. |
| [26] | EVANS L M, SLAVOV G T, RODGERS-MELNICK E, et al. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations[J]. Nature Genetics, 2014, 46(10): 1089-1096. |
| [27] | XUE L J, WU H T, CHEN Y N, et al. Evidences for a role of two Y-specific genes in sex determination in Populus deltoides[J]. Nature Communications, 2020, 11: 5893. |
| [28] | BAI S J, WU H N, ZHANG J P, et al. Genome assembly of salicaceae Populus deltoides(eastern cottonwood) I-69 based on nanopore sequencing and Hi-C technologies[J]. Journal of Heredity, 2021, 112(3): 303-310. |
| [29] | QIU D Y, BAI S L, MA J C, et al. The genome of Populus alba×Populus tremula var. glandulosa clone 84K[J]. DNA Research, 2019, 26(5): 423-431. |
| [30] | AN X M, GAO K, CHEN Z, et al. High quality haplotype-resolved genome assemblies of Populus tomentosa Carr., a stabilized interspecific hybrid species widespread in Asia[J]. Molecular Ecology Resources, 2022, 22(2): 786-802. |
| [31] | TONG S F, WANG Y B, CHEN N N, et al. PtoNF-YC9-SRMT-PtoRD26 module regulates the high saline tolerance of a triploid poplar[J]. Genome Biology, 2022, 23(1): 148. |
| [32] | 李建国, 濮励杰, 朱明, 等. 土壤盐渍化研究现状及未来研究热点[J]. 地理学报, 2012, 67(9): 1233-1245. |
| LI J G, PU L J, ZHU M, et al. The present situation and hot issues in the salt-affected soil research[J]. Acta Geographica Sinica, 2012, 67(9): 1233-1245. (in Chinese with English abstract) | |
| [33] | 储陈辰. 杨属和柳属泛基因组构建与基因组变异分析[D]. 南京: 南京林业大学, 2023. |
| CHU C C. Pan-genome and genomic variation analysis of genera Populus and Salix[D]. Nanjing: Nanjing Forestry University, 2023. (in Chinese with English abstract) | |
| [34] | XU Z S, NI Z Y, LIU L, et al. Characterization of the TaAIDFa gene encoding a CRT/DRE-binding factor responsive to drought, high-salt, and cold stress in wheat[J]. Molecular Genetics and Genomics, 2008, 280(6): 497-508. |
| [35] | ERPEN L, DEVI H S, GROSSER J W, et al. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants[J]. Plant Cell, Tissue and Organ Culture, 2018, 132(1): 1-25. |
| [36] | HENDERSON S W, DUNLEVY J D, WU Y, et al. Functional differences in transport properties of natural HKT1;1 variants influence shoot Na+ exclusion in grapevine rootstocks[J]. New Phytologist, 2018, 217(3): 1113-1127. |
| [37] | WU Y, HENDERSON S W, WEGE S, et al. The grapevine NaE sodium exclusion locus encodes sodium transporters with diverse transport properties and localisation[J]. Journal of Plant Physiology, 2020, 246: 153113. |
| [38] | ZHANG M, CAO Y B, WANG Z P, et al. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize[J]. New Phytologist, 2018, 217(3): 1161-1176. |
| [39] | ALNAYEF M, SOLIS C, SHABALA L, et al. Changes in expression level of OsHKT1;5 alters activity of membrane transporters involved in K+ and Ca2+ acquisition and homeostasis in salinized rice roots[J]. International Journal of Molecular Sciences, 2020, 21(14): 4882. |
| [1] | HU Yingjie, DU Chenqi, WANG Liufan, SHOU Jianxin, WANG Chao, XU Mei, YAN Xu. Research progress of vesicle trafficking in plant response to salt stress [J]. Acta Agriculturae Zhejiangensis, 2025, 37(9): 2003-2011. |
| [2] | GUAN Xiusheng, LIU Tieshan, WANG Juan, ZHANG Maolin, LIU Chunxiao, DONG Rui, GUAN Haiying, LIU Qiang, XU Yang, HE Chunmei. Bioinformatics analysis and cloning of NF-YA family genes in maize(Zea mays) [J]. Acta Agriculturae Zhejiangensis, 2025, 37(8): 1605-1614. |
| [3] | WU Guojiang, ZHOU Wei, LI Yanxiao, HOU Jie, YANG Zhiqiang, ZHOU Yaxing. Identification and expression analysis under saline-alkali stress of ZF-HD gene family in sorghum [J]. Acta Agriculturae Zhejiangensis, 2024, 36(6): 1217-1231. |
| [4] | LI Yaping, JIN Fulai, HUANG Zonggui, ZHANG Tao, DUAN Xiaojing, JIANG Wu, TAO Zhengming, CHEN Jiadong. Identification and expression pattern analysis of glycoside hydrolase GH3 gene family in Dendrobium officinale [J]. Acta Agriculturae Zhejiangensis, 2024, 36(4): 790-799. |
| [5] | GAO Jing, LU Linghong, GU Xianbin, FAN Fei, SONG Genhua, ZHANG Huiqin. Cloning of AcWRKY94 gene from kiwifruit and its functional analysis under salt stress [J]. Acta Agriculturae Zhejiangensis, 2024, 36(11): 2501-2509. |
| [6] | TANG Yuehui, CHEN Shuying, HE Wenqiong, WANG Hanjin, BAO Xinxin, JIA Sainan, WANG Yaoyao, CHEN Yuyang, YANG Tongwen. Cloning and functional analysis of JcERF22 gene from Jatropha curcas [J]. Acta Agriculturae Zhejiangensis, 2024, 36(10): 2219-2228. |
| [7] | MA Zhonghua, WU Na, CHEN Juan, ZHAO Cong, YAN Chenghong, LIU Jili. Effects of salt stress and phosphorus supply on physiological characteristics of switchgrass seedlings [J]. Acta Agriculturae Zhejiangensis, 2022, 34(6): 1205-1216. |
| [8] | LI Liyan, TAN Haixia, LI Jing, WANG Lianlong, DU Yinghui, XU Zhiwen. Screening of salt-tolerant growth-promoting Bacillus strains and their effect on oat growth under salt stress [J]. Acta Agriculturae Zhejiangensis, 2022, 34(6): 1268-1276. |
| [9] | LIU Chen, XU Haobo, SI Yuyang, LI Yapeng, GUO Yuting, DU Changxia. Research progress on regulation mechanism of plant response to salt stress based on transcriptomics [J]. Acta Agriculturae Zhejiangensis, 2022, 34(4): 870-878. |
| [10] | YANG Xinxia, TANG Mansheng, ZHANG Bin. Identification of soybean PP2C family genes and transcriptome analysis in response to salt stress [J]. Acta Agriculturae Zhejiangensis, 2022, 34(2): 207-220. |
| [11] | LIU Tao, CHEN Hairong, WANG Chengzhong, REN Li, ZHANG Di. Physiology of stress resistance of Agapanthus praecox under drought and salt stress [J]. Acta Agriculturae Zhejiangensis, 2022, 34(12): 2669-2681. |
| [12] | MENG Na, XUE Hui, WEI Ming, WEI Shenghua. Ion characteristics on chloride channel blocker ameliorating salt injury to Glycine max [J]. Acta Agriculturae Zhejiangensis, 2022, 34(10): 2095-2104. |
| [13] | ZHOU Beining, MAO Lian, HUA Zhuangzhuang, LU Jianguo. Effects of alkaline salt stress on growth and ion allocation of Sinocalycanthus chinensis [J]. Acta Agriculturae Zhejiangensis, 2022, 34(1): 79-88. |
| [14] | YANG Xinxia, ZHANG Bin. Identification of soybean LAZ1 gene family and functional analysis of GmLAZ1-9 [J]. Acta Agriculturae Zhejiangensis, 2021, 33(4): 586-594. |
| [15] | LU Anqiao, ZHANG Fengju, WANG Xueqin, XU Xing. Effects of NaCl and Na2SO4 stress on content and distribution of K + and Na + of Echinochloa frumentacea seedlings [J]. Acta Agriculturae Zhejiangensis, 2021, 33(3): 396-403. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||